Rapid and sensitive large-scale screening of low affinity extracellular receptor protein interactions by using reaction induced inhibition of Gaussia luciferase
- PMID: 32601498
- PMCID: PMC7324543
- DOI: 10.1038/s41598-020-67468-7
Rapid and sensitive large-scale screening of low affinity extracellular receptor protein interactions by using reaction induced inhibition of Gaussia luciferase
Abstract
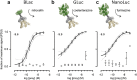
Extracellular protein interactions mediated by cell surface receptors are essential for intercellular communication in multicellular organisms. Assays to detect extracellular interactions must account for their often weak binding affinities and also the biochemical challenges in solubilising membrane-embedded receptors in an active form. Methods based on detecting direct binding of soluble recombinant receptor ectodomains have been successful, but genome-scale screening is limited by the usual requirement of producing sufficient amounts of each protein in two different forms, usually a "bait" and "prey". Here, we show that oligomeric receptor ectodomains coupled to concatenated units of the light-generating Gaussia luciferase enzyme robustly detected low affinity interactions and reduced the amount of protein required by several orders of magnitude compared to other reporter enzymes. Importantly, we discovered that this flash-type luciferase exhibited a reaction-induced inhibition that permitted the use of a single protein preparation as both bait and prey thereby halving the number of expression plasmids and recombinant proteins required for screening. This approach was tested against a benchmarked set of quantified extracellular interactions and shown to detect extremely weak interactions (KDs ≥ μM). This method will facilitate large-scale receptor interaction screening and contribute to the goal of mapping networks of cellular communication.
Conflict of interest statement
The authors declare no competing interests.
Figures

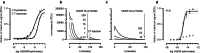

References
-
- Ben-Shlomo, I., Yu Hsu, S., Rauch, R., Kowalski, H. W. & Hsueh, A. J. Signaling receptome: a genomic and evolutionary perspective of plasma membrane receptors involved in signal transduction. Sci. STKE2003, RE9, doi:10.1126/stke.2003.187.re9 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

