Large-scale pathway specific polygenic risk and transcriptomic community network analysis identifies novel functional pathways in Parkinson disease
- PMID: 32601912
- PMCID: PMC8096770
- DOI: 10.1007/s00401-020-02181-3
Large-scale pathway specific polygenic risk and transcriptomic community network analysis identifies novel functional pathways in Parkinson disease
Erratum in
-
Correction to: Large‑scale pathway specific polygenic risk and transcriptomic community network analysis identifies novel functional pathways in Parkinson disease.Acta Neuropathol. 2021 Jul;142(1):223-224. doi: 10.1007/s00401-021-02309-z. Acta Neuropathol. 2021. PMID: 33944973 Free PMC article. No abstract available.
Abstract
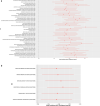
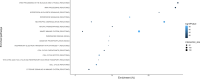
Polygenic inheritance plays a central role in Parkinson disease (PD). A priority in elucidating PD etiology lies in defining the biological basis of genetic risk. Unraveling how risk leads to disruption will yield disease-modifying therapeutic targets that may be effective. Here, we utilized a high-throughput and hypothesis-free approach to determine biological processes underlying PD using the largest currently available cohorts of genetic and gene expression data from International Parkinson's Disease Genetics Consortium (IPDGC) and the Accelerating Medicines Partnership-Parkinson's disease initiative (AMP-PD), among other sources. We applied large-scale gene-set specific polygenic risk score (PRS) analyses to assess the role of common variation on PD risk focusing on publicly annotated gene sets representative of curated pathways. We nominated specific molecular sub-processes underlying protein misfolding and aggregation, post-translational protein modification, immune response, membrane and intracellular trafficking, lipid and vitamin metabolism, synaptic transmission, endosomal-lysosomal dysfunction, chromatin remodeling and apoptosis mediated by caspases among the main contributors to PD etiology. We assessed the impact of rare variation on PD risk in an independent cohort of whole-genome sequencing data and found evidence for a burden of rare damaging alleles in a range of processes, including neuronal transmission-related pathways and immune response. We explored enrichment linked to expression cell specificity patterns using single-cell gene expression data and demonstrated a significant risk pattern for dopaminergic neurons, serotonergic neurons, hypothalamic GABAergic neurons, and neural progenitors. Subsequently, we created a novel way of building de novo pathways by constructing a network expression community map using transcriptomic data derived from the blood of PD patients, which revealed functional enrichment in inflammatory signaling pathways, cell death machinery related processes, and dysregulation of mitochondrial homeostasis. Our analyses highlight several specific promising pathways and genes for functional prioritization and provide a cellular context in which such work should be done.
Keywords: Mendelian randomization; Parkinson disease; Polygenic risk; Transcriptome community maps.
Conflict of interest statement
No other disclosures were reported.
Figures


References
-
- Combe D, Largeron C, Géry M, Egyed-Zsigmond E. I-Louvain: an attributed graph clustering method. In: Fromont E, De Bie T, van Leeuwen M, editors. Advances in intelligent data analysis. Cham: Springer; 2015.
-
- Centers for Common Disease Genomics. In: Genome.gov. https://www.genome.gov/Funded-Programs-Projects/NHGRI-Genome-Sequencing-.... Accessed 12 Nov 2019
Publication types
MeSH terms
Grants and funding
- G0802462/MRC_/Medical Research Council/United Kingdom
- Z01 AG000949/ImNIH/Intramural NIH HHS/United States
- U01 NS100603/NS/NINDS NIH HHS/United States
- UL1 RR025774/RR/NCRR NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- MR/N008324/1/MRC_/Medical Research Council/United Kingdom
- R01 NS115144/NS/NINDS NIH HHS/United States
- MR/K01417X/1/MRC_/Medical Research Council/United Kingdom
- Z99 AG999999/ImNIH/Intramural NIH HHS/United States
- Z01-AG000949-0/AG/NIA NIH HHS/United States
- Z01 ES101986/ImNIH/Intramural NIH HHS/United States
- U24 NS095871/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

