PharmVar GeneFocus: CYP2C19
- PMID: 32602114
- PMCID: PMC7769975
- DOI: 10.1002/cpt.1973
PharmVar GeneFocus: CYP2C19
Abstract
The Pharmacogene Variation Consortium (PharmVar) catalogues star (*) allele nomenclature for the polymorphic human CYP2C19 gene. CYP2C19 genetic variation impacts the metabolism of many drugs and has been associated with both efficacy and safety issues for several commonly prescribed medications. This GeneFocus provides a comprehensive overview and summary of CYP2C19 and describes how haplotype information catalogued by PharmVar is utilized by the Pharmacogenomics Knowledgebase and the Clinical Pharmacogenetics Implementation Consortium (CPIC).
© 2020 The Author. Clinical Pharmacology & Therapeutics published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
Conflicts of Interest:
Indiana University School of Medicine Pharmacogenomics Laboratory, Millennium Health, and Sema4 are fee-for-service clinical laboratories that offer clinical pharmacogenetic testing. A.L.D. is a paid employee of Millennium Health; S.A.S., is a paid employee of Sema4. All other authors declared no competing interests for this work.
Figures
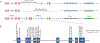

 )symbol indicates that a core SNV alters function and the PharmVar (
)symbol indicates that a core SNV alters function and the PharmVar ( ) symbol highlights that a core SNV is unique to a star allele. SNV positions refer to genomic coordinates on the NG_008384.3 reference sequence (with the ATG start codon being +1).
) symbol highlights that a core SNV is unique to a star allele. SNV positions refer to genomic coordinates on the NG_008384.3 reference sequence (with the ATG start codon being +1).
References
-
- Kupfer A & Preisig R Pharmacogenetics of mephenytoin: a new drug hydroxylation polymorphism in man. Eur J Clin Pharmacol 26, 753–9 (1984). - PubMed
-
- Goldstein JA et al. Evidence that CYP2C19 is the major (S)-mephenytoin 4’-hydroxylase in humans. Biochemistry 33, 1743–52 (1994). - PubMed
-
- Wrighton SA, Stevens JC, Becker GW & VandenBranden M Isolation and characterization of human liver cytochrome P450 2C19: correlation between 2C19 and S-mephenytoin 4’-hydroxylation. Arch Biochem Biophys 306, 240–5 (1993). - PubMed
-
- Romkes M, Faletto MB, Blaisdell JA, Raucy JL & Goldstein JA Cloning and expression of complementary DNAs for multiple members of the human cytochrome P450IIC subfamily. Biochemistry 30, 3247–55 (1991). - PubMed
-
- Goldstein JA & de Morais SM Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics 4, 285–99 (1994). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

