Sensitive and broadly applicable residual disease detection in acute myeloid leukemia using flow cytometry-based leukemic cell enrichment followed by mutational profiling
- PMID: 32602117
- PMCID: PMC7540028
- DOI: 10.1002/ajh.25918
Sensitive and broadly applicable residual disease detection in acute myeloid leukemia using flow cytometry-based leukemic cell enrichment followed by mutational profiling
Abstract
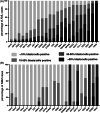
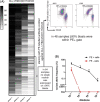
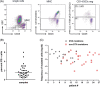
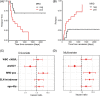
Persistent measurable residual disease (MRD) is an increasingly important prognostic marker in acute myeloid leukemia (AML). Currently, MRD is determined by multi-parameter flow cytometry (MFC) or PCR-based methods detecting leukemia-specific fusion transcripts and mutations. However, while MFC is highly operator-dependent and difficult to standardize, PCR-based methods are only available for a minority of AML patients. Here we describe a novel, highly sensitive and broadly applicable method for MRD detection by combining MFC-based leukemic cell enrichment using an optimized combinatorial antibody panel targeting CLL-1, TIM-3, CD123 and CD117, followed by mutational analysis of recurrently mutated genes in AML. In dilution experiments this method showed a sensitivity of 10-4 to 10-5 for residual disease detection. In prospectively collected remission samples this marker combination allowed for a median 67-fold cell enrichment with sufficient DNA quality for mutational analysis using next generation sequencing (NGS) or digital PCR in 39 out of 41 patients. Twenty-one samples (53.8%) tested MRD positive, whereas 18 (46.2%) were negative. With a median follow-up of 559 days, 71.4% of MRD positive (15/21) and 27.8% (5/18) of MRD negative patients relapsed (P = .007). The cumulative incidence of relapse (CIR) was higher for MRD positive patients (5-year CIR: 90.5% vs 28%, P < .001). In multivariate analysis, MRD positivity was a prominent factor for CIR. Thus, MFC-based leukemic cell enrichment using antibodies against CLL-1, TIM-3, CD123 and CD117 followed by mutational analysis allows high sensitive MRD detection and is informative on relapse risk in the majority of AML patients.
© 2020 The Authors. American Journal of Hematology published by Wiley Periodicals LLC.
Conflict of interest statement
B.P. and A.W. report research support from Becton Dickinson BioSciences. All other authors declare no conflict of interest.
Figures
References
-
- Bachas C, Schuurhuis GJ, Assaraf YG, et al. The role of minor subpopulations within the leukemic blast compartment of AML patients at initial diagnosis in the development of relapse. Leukemia. 2012;26:1313‐1320. - PubMed
-
- Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136‐1152. - PubMed
-
- Yin JA, O'Brien MA, Hills RK, Daly SB, Wheatley K, Burnett AK. Minimal residual disease monitoring by quantitative RT‐PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML‐15 trial. Blood. 2012;120:2826‐2835. - PubMed
-
- Ivey A, Hills RK, Simpson MA, et al. Assessment of minimal residual disease in standard‐risk AML. N Engl J Med. 2016;374(5):422‐433. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
