DNA metabarcoding effectively quantifies diatom responses to nutrients in streams
- PMID: 32602216
- PMCID: PMC7731896
- DOI: 10.1002/eap.2205
DNA metabarcoding effectively quantifies diatom responses to nutrients in streams
Abstract
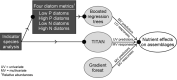
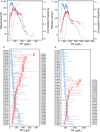
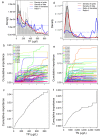
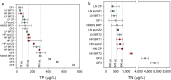
Nutrient pollution from human activities remains a common problem facing stream ecosystems. Identifying ecological responses to phosphorus and nitrogen can inform decisions affecting the protection and management of streams and their watersheds. Diatoms are particularly useful because they are a highly diverse group of unicellular algae found in nearly all aquatic environments and are sensitive responders to increased nutrient concentrations. Here, we used DNA metabarcoding of stream diatoms as an approach to quantifying effects of total phosphorus (TP) and total nitrogen (TN). Threshold indicator taxa analysis (TITAN) identified operational taxonomic units (OTUs) that increased or decreased along TP and TN gradients along with nutrient concentrations at which assemblages had substantial changes in the occurrences and relative abundances of OTUs. Boosted regression trees showed that relative abundances of gene sequence reads for OTUs identified by TITAN as low P, high P, low N, or high N diatoms had strong relationships with nutrient concentrations, which provided support for potentially using these groups of diatoms as metrics in monitoring programs. Gradient forest analysis provided complementary information by characterizing multi-taxa assemblage change using multiple predictors and results from random forest models for each OTU. Collectively, these analyses showed that notable changes in diatom assemblage structure and OTUs began around 20 µg TP/L, low P diatoms decreased substantially and community change points occurred from 75 to 150 µg/L, and high P diatoms became increasingly dominant from 150 to 300 µg/L. Diatoms also responded to TN with large decreases in low N diatoms occurring from 280 to 525 µg TN/L and a transition to dominance by high N diatoms from 525-850 µg/L. These diatom responses to TP and TN could be used to inform protection efforts (i.e., anti-degradation) and management goals (i.e., nutrient reduction) in streams and watersheds. Our results add to the growing support for using diatom metabarcoding in monitoring programs.
Keywords: agriculture; algae; bioassessment; biomonitoring; boosted regression trees; gradient forest; nitrogen; periphyton; phosphorus; rbcL; threshold indicator taxa analysis.
Published 2020. This article is a U.S. Government work and is in the public domain in the USA. Ecological Applications published by Wiley Periodicals LLC on behalf of Ecological Society of America.
Figures


References
-
- Apothéloz‐Perret‐Gentil, L. , Cordonier A., Straub F., Iseli J., Esling P., and Pawlowski J.. 2017. Taxonomy‐free molecular diatom index for high‐throughput eDNA biomonitoring. Molecular Ecology Resources 17:1231–1242. - PubMed
-
- Baker, M. E. , and King R. S.. 2010. A new method for detecting and interpreting biodiversity and ecological community thresholds. Methods in Ecology and Evolution 1:25–37.
-
- Baker, M. E. , King R. S., and Kahle D..2015. TITAN2: Threshold Indicator Taxa Analysis. https://cran.r‐project.org/web/packages/TITAN2/TITAN2.pdf
-
- Becker, M. E. , Becker T. J., and Bellucci C. J.. 2019. Diatom tolerance metrics to identify total phosphorus as candidate cause of aquatic life impairment in Connecticut, USA freshwater streams. Ecological Indicators 93:638–646.
-
- Breiman, L. 2001. Random forests. Machine Learning 45:5–32.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

