Targeted surveillance strategies for efficient detection of novel antibiotic resistance variants
- PMID: 32602459
- PMCID: PMC7326491
- DOI: 10.7554/eLife.56367
Targeted surveillance strategies for efficient detection of novel antibiotic resistance variants
Abstract
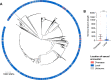
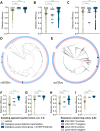
Genotype-based diagnostics for antibiotic resistance represent a promising alternative to empiric therapy, reducing inappropriate antibiotic use. However, because such assays infer resistance based on known genetic markers, their utility will wane with the emergence of novel resistance. Maintenance of these diagnostics will therefore require surveillance to ensure early detection of novel resistance variants, but efficient strategies to do so remain undefined. We evaluate the efficiency of targeted sampling approaches informed by patient and pathogen characteristics in detecting antibiotic resistance and diagnostic escape variants in Neisseria gonorrhoeae, a pathogen associated with a high burden of disease and antibiotic resistance and the development of genotype-based diagnostics. We show that patient characteristic-informed sampling is not a reliable strategy for efficient variant detection. In contrast, sampling informed by pathogen characteristics, such as genomic diversity and genomic background, is significantly more efficient than random sampling in identifying genetic variants associated with resistance and diagnostic escape.
Keywords: Neisseria gonorrhoeae; antibiotic resistance; diagnostic; epidemiology; global health; infectious disease; microbiology; surveillance.
© 2020, Hicks et al.
Conflict of interest statement
AH, SK, TM, KM, GT, MA, DW, YG No competing interests declared, ML Reviewing editor, eLife
Figures



Comment in
-
Genomics against gonorrhoea.Elife. 2020 Jun 30;9:e59379. doi: 10.7554/eLife.59379. Elife. 2020. PMID: 32602460 Free PMC article.
References
-
- André E, Goeminne L, Colmant A, Beckert P, Niemann S, Delmee M. Novel rapid PCR for the detection of Ile491Phe rpoB mutation of Mycobacterium tuberculosis , a rifampicin-resistance-conferring mutation undetected by commercial assays. Clinical Microbiology and Infection. 2017;23:26267. doi: 10.1016/j.cmi.2016.12.009. - DOI - PubMed
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
-
- Bazzo ML, Golfetto L, Gaspar PC, Pires AF, Ramos MC, Franchini M, Ferreira WA, Unemo M, Benzaken AS, Brazilian-GASP Network First nationwide antimicrobial susceptibility surveillance for Neisseria gonorrhoeae in Brazil, 2015-16. Journal of Antimicrobial Chemotherapy. 2018;73:1854–1861. doi: 10.1093/jac/dky090. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

