Quantitative proteomics analysis of young and elderly skin with DIA mass spectrometry reveals new skin aging-related proteins
- PMID: 32602849
- PMCID: PMC7377841
- DOI: 10.18632/aging.103461
Quantitative proteomics analysis of young and elderly skin with DIA mass spectrometry reveals new skin aging-related proteins
Abstract
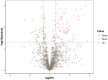
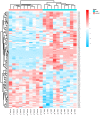
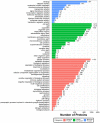
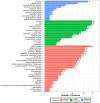
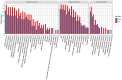
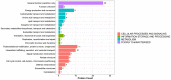
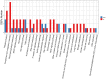
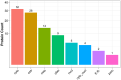
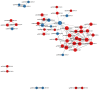
Skin aging is a specific manifestation of the physiological aging process that occurs in virtually all organisms. In this study, we used data independent acquisition mass spectrometry to perform a comparative analysis of protein expression in volar forearm skin samples from of 20 healthy young and elderly Chinese individuals. Our quantitative proteomic analysis identified a total of 95 differentially expressed proteins (DEPs) in aged skin compared to young skin. Enrichment analyses of these DEPs (57 upregulated and 38 downregulated proteins) based on the GO, KEGG, and KOG databases revealed functional clusters associated with immunity and inflammation, oxidative stress, biosynthesis and metabolism, proteases, cell proliferation, cell differentiation, and apoptosis. We also found that GAPDH, which was downregulated in aged skin samples, was the top hub gene in a protein-protein interaction network analysis. Some of the DEPs identified herein had been previously correlated with aging of the skin and other organs, while others may represent novel age-related entities. Our non-invasive proteomics analysis of human epidermal proteins may guide future research on skin aging to help develop treatments for age-related skin conditions and rejuvenation.
Keywords: aging; epidermal proteins; mass spectrometer; proteome; skin rejuvenation and aging.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

