Age-of-onset information helps identify 76 genetic variants associated with allergic disease
- PMID: 32603359
- PMCID: PMC7367489
- DOI: 10.1371/journal.pgen.1008725
Age-of-onset information helps identify 76 genetic variants associated with allergic disease
Abstract
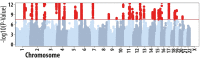
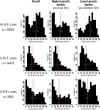
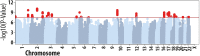
Risk factors that contribute to inter-individual differences in the age-of-onset of allergic diseases are poorly understood. The aim of this study was to identify genetic risk variants associated with the age at which symptoms of allergic disease first develop, considering information from asthma, hay fever and eczema. Self-reported age-of-onset information was available for 117,130 genotyped individuals of European ancestry from the UK Biobank study. For each individual, we identified the earliest age at which asthma, hay fever and/or eczema was first diagnosed and performed a genome-wide association study (GWAS) of this combined age-of-onset phenotype. We identified 50 variants with a significant independent association (P<3x10-8) with age-of-onset. Forty-five variants had comparable effects on the onset of the three individual diseases and 38 were also associated with allergic disease case-control status in an independent study (n = 222,484). We observed a strong negative genetic correlation between age-of-onset and case-control status of allergic disease (rg = -0.63, P = 4.5x10-61), indicating that cases with early disease onset have a greater burden of allergy risk alleles than those with late disease onset. Subsequently, a multivariate GWAS of age-of-onset and case-control status identified a further 26 associations that were missed by the univariate analyses of age-of-onset or case-control status only. Collectively, of the 76 variants identified, 18 represent novel associations for allergic disease. We identified 81 likely target genes of the 76 associated variants based on information from expression quantitative trait loci (eQTL) and non-synonymous variants, of which we highlight ADAM15, FOSL2, TRIM8, BMPR2, CD200R1, PRKCQ, NOD2, SMAD4, ABCA7 and UBE2L3. Our results support the notion that early and late onset allergic disease have partly distinct genetic architectures, potentially explaining known differences in pathophysiology between individuals.
Conflict of interest statement
C. Tian and D. A. Hinds both report support from 23andMe during the conduct of the study. C. Almqvist received grant 2017-00641 from Swedish Research Council and Swedish Initiative for Research on Microdata in the Social And Medical Sciences (SIMSAM) framework grant (340-2013-5867) for this work. G. H. Koppelman's institution received grants from the Lung Foundation of The Netherlands, the Ubbo Emmius Foundation, TEVA (The Netherlands), GlaxoSmithKline, Vertex, and the Tetri Foundation for other works. L. Paternoster received grant MR/J012165/1 from the UK Medical Research Council for this work and personal fees from Merck. The rest of the authors declare that they have no relevant conflicts of interest.
Figures