Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome
- PMID: 32603407
- PMCID: PMC7537389
- DOI: 10.1084/jem.20200483
Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome
Abstract
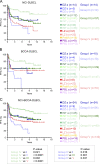
In response to T cell-dependent antigens, mature B cells are stimulated to form germinal centers (GCs), the sites of B cell affinity maturation and the cell of origin (COO) of most B cell lymphomas. To explore the dynamics of GC B cell development beyond the known dark zone and light zone compartments, we performed single-cell (sc) transcriptomic analysis on human GC B cells and identified multiple functionally linked subpopulations, including the distinct precursors of memory B cells and plasma cells. The gene expression signatures associated with these GC subpopulations were effective in providing a sc-COO for ∼80% of diffuse large B cell lymphomas (DLBCLs) and identified novel prognostic subgroups of DLBCL.
© 2020 Holmes et al.
Conflict of interest statement
Disclosures: N. Compagno is currently employed at Novartis. No other disclosures were reported.
Figures

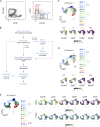




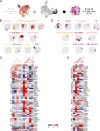


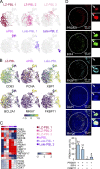

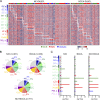


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

