Evaluation of Peptide-Based Probes toward In Vivo Diagnostic Imaging of Bacterial Biofilm-Associated Infections
- PMID: 32603591
- PMCID: PMC7429274
- DOI: 10.1021/acsinfecdis.0c00125
Evaluation of Peptide-Based Probes toward In Vivo Diagnostic Imaging of Bacterial Biofilm-Associated Infections
Abstract
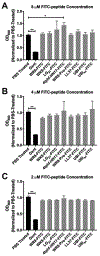
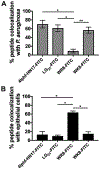
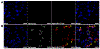
The clinical management of bacterial biofilm infections represents an enormous challenge in today's healthcare setting. The NIH estimates that 65% of bacterial infections are biofilm-related, and therapeutic outcomes are positively correlated with early intervention. Currently, there is no reliable imaging technique to detect biofilm infections in vivo, and current clinical protocols for accurate and direct biofilm identification are nonexistent. In orthopedic implant-associated biofilm infections, for example, current detection methods are based on nonspecific X-ray or radiolabeled white blood cell imaging, coupled with peri-prosthetic tissue or fluid samples taken invasively, and must be cultured. This approach is time-consuming and often fails to detect biofilm bacteria due to sampling errors and a lack of sensitivity. The ability to quantify bacterial biofilms by real-time noninvasive imaging is an urgent unmet clinical need that would revolutionize the management and treatment of these devastating types of infections. In the present study, we assembled a collection of fluorescently labeled peptide candidates to specifically explore their biofilm targeting properties. We evaluated these fluorescently labeled peptides using various in vitro assays for their ability to specifically and nondestructively target biofilms produced by model bacterial pathogen Pseudomonas aeruginosa. The lead candidate that emerged, 4Iphf-HN17, demonstrated rapid biofilm labeling kinetics, a lack of bactericidal activity, and biofilm targeting specificity in human cell infection models. In vivo fluorescently labeled 4Iphf-HN17 showed enhanced accumulation in biofilm-infected wounds, thus warranting further study.
Keywords: Pseudomonas aeruginosa; biofilms; diagnostics; imaging; optical; peptides.
Figures





References
-
- Davies D, Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2003, 2 (2), 114–22. - PubMed
-
- Bjarnsholt T; Alhede M; Alhede M; Eickhardt-Sorensen SR; Moser C; Kuhl M; Jensen PO; Hoiby N, The in vivo biofilm. Trends Microbiol 2013, 21 (9), 466–74. - PubMed
-
- Flemming HC; Wingender J, The biofilm matrix. Nat Rev Microbiol 2010, 8 (9), 623–33. - PubMed
-
- Bjarnsholt T, The role of bacterial biofilms in chronic infections. APMIS Suppl 2013, (136), 1–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

