Single-Cell Transcriptional Archetypes of Airway Inflammation in Cystic Fibrosis
- PMID: 32603604
- PMCID: PMC7667912
- DOI: 10.1164/rccm.202004-0991OC
Single-Cell Transcriptional Archetypes of Airway Inflammation in Cystic Fibrosis
Abstract
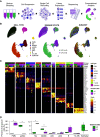
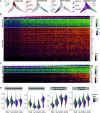
Rationale: Cystic fibrosis (CF) is a life-shortening, multisystem hereditary disease caused by abnormal chloride transport. CF lung disease is driven by innate immune dysfunction and exaggerated inflammatory responses that contribute to tissue injury. To define the transcriptional profile of this airway immune dysfunction, we performed the first single-cell transcriptome characterization of CF sputum.Objectives: To define the transcriptional profile of sputum cells and its implication in the pathogenesis of immune function and the development of CF lung disease.Methods: We performed single-cell RNA sequencing of sputum cells from nine subjects with CF and five healthy control subjects. We applied novel computational approaches to define expression-based cell function and maturity profiles, herein called transcriptional archetypes.Measurements and Main Results: The airway immune cell repertoire shifted from alveolar macrophages in healthy control subjects to a predominance of recruited monocytes and neutrophils in CF. Recruited lung mononuclear phagocytes were abundant in CF and were separated into the following three archetypes: activated monocytes, monocyte-derived macrophages, and heat shock-activated monocytes. Neutrophils were the most prevalent in CF, with a dominant immature proinflammatory archetype. Although CF monocytes exhibited proinflammatory features, both monocytes and neutrophils showed transcriptional evidence of abnormal phagocytic and cell-survival programs.Conclusions: Our findings offer an opportunity to understand subject-specific immune dysfunction and its contribution to divergent clinical courses in CF. As we progress toward personalized applications of therapeutic and genomic developments, we hope this inflammation-profiling approach will enable further discoveries that change the natural history of CF lung disease.
Keywords: RNA-seq; cystic fibrosis; macrophages; monocytes; neutrophils.
Figures

Comment in
-
Spelunking in Sputum: Single-Cell RNA Sequencing Sheds New Insights into Cystic Fibrosis.Am J Respir Crit Care Med. 2020 Nov 15;202(10):1336-1337. doi: 10.1164/rccm.202006-2456ED. Am J Respir Crit Care Med. 2020. PMID: 32721178 Free PMC article. No abstract available.
References
-
- Cystic Fibrosis Foundation. Cystic Fibrosis Foundation patient registry 2017 annual data report. 2018 [2020 Jan 15] Available from: https://www.cff.org/Research/Researcher-Resources/Patient-Registry/2017-....
-
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. - PubMed
-
- Welsh MJ, Ramsey BW, Accurso F, Cutting GR. Cystic fibrosis. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Vogelstein B, editors. The metabolic and molecular basis of inherited disease. New York: McGraw-Hill; 2001. pp. 5121–5189.
Publication types
MeSH terms
Grants and funding
- K08 HL135402/HL/NHLBI NIH HHS/United States
- R21 LM012884/LM/NLM NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- T32 HL007778/HL/NHLBI NIH HHS/United States
- R01 HL153604/HL/NHLBI NIH HHS/United States
- U01 HL145567/HL/NHLBI NIH HHS/United States
- K01 HL125514/HL/NHLBI NIH HHS/United States
- R01 HL127349/HL/NHLBI NIH HHS/United States
- R03 HL154275/HL/NHLBI NIH HHS/United States
- K01 HL125474/HL/NHLBI NIH HHS/United States
- R03 HL154276/HL/NHLBI NIH HHS/United States
- R01 HL155948/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

