Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo'
- PMID: 32604404
- PMCID: PMC7618354
- DOI: 10.1038/s41586-020-2488-1
Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo'
Erratum in
-
Author Correction: Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo'.Nature. 2021 Feb;590(7844):E11. doi: 10.1038/s41586-020-2956-7. Nature. 2021. PMID: 33452443 Free PMC article. No abstract available.
Abstract
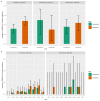
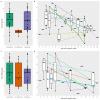
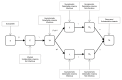
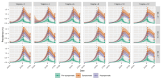
On 21 February 2020, a resident of the municipality of Vo', a small town near Padua (Italy), died of pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection1. This was the first coronavirus disease 19 (COVID-19)-related death detected in Italy since the detection of SARS-CoV-2 in the Chinese city of Wuhan, Hubei province2. In response, the regional authorities imposed the lockdown of the whole municipality for 14 days3. Here we collected information on the demography, clinical presentation, hospitalization, contact network and the presence of SARS-CoV-2 infection in nasopharyngeal swabs for 85.9% and 71.5% of the population of Vo' at two consecutive time points. From the first survey, which was conducted around the time the town lockdown started, we found a prevalence of infection of 2.6% (95% confidence interval (CI): 2.1-3.3%). From the second survey, which was conducted at the end of the lockdown, we found a prevalence of 1.2% (95% CI: 0.8-1.8%). Notably, 42.5% (95% CI: 31.5-54.6%) of the confirmed SARS-CoV-2 infections detected across the two surveys were asymptomatic (that is, did not have symptoms at the time of swab testing and did not develop symptoms afterwards). The mean serial interval was 7.2 days (95% CI: 5.9-9.6). We found no statistically significant difference in the viral load of symptomatic versus asymptomatic infections (P = 0.62 and 0.74 for E and RdRp genes, respectively, exact Wilcoxon-Mann-Whitney test). This study sheds light on the frequency of asymptomatic SARS-CoV-2 infection, their infectivity (as measured by the viral load) and provides insights into its transmission dynamics and the efficacy of the implemented control measures.
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Italian Vò municipality cohort and COVID-19 epidemiology: The "Framingham" study of the 21st century.Eur J Intern Med. 2020 Nov;81:95-96. doi: 10.1016/j.ejim.2020.07.015. Epub 2020 Jul 21. Eur J Intern Med. 2020. PMID: 32709547 Free PMC article. No abstract available.
References
-
- Crisanti A, Cassone A. In one Italian town, we showed mass testing could eradicate the coronavirus. The Guardian. 2020 https://www.theguardian.com/commentisfree/2020/mar/20/eradicated-coronav... .
-
- World Health Organization. Novel coronavirus (COVID-19) situation. WHO; 2020. https://covid19.who.int/
-
- Saini V. Coronavirus: voices from a quarantined Italian town. EU Observer. 2020 https://euobserver.com/coronavirus/147552 .
-
- Volz E, et al. Report 5: phylogenetic analysis of SARS-CoV-2. Imperial College London; 2020. https://www.imperial.ac.uk/media/imperial-college/medicine/mrc-gida/2020... .
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

