Metabolic Activation of CsgD in the Regulation of Salmonella Biofilms
- PMID: 32604994
- PMCID: PMC7409106
- DOI: 10.3390/microorganisms8070964
Metabolic Activation of CsgD in the Regulation of Salmonella Biofilms
Abstract
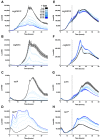
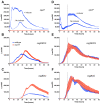
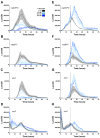
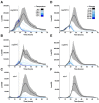
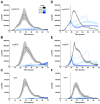
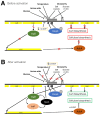
Among human food-borne pathogens, gastroenteritis-causing Salmonella strains have the most real-world impact. Like all pathogens, their success relies on efficient transmission. Biofilm formation, a specialized physiology characterized by multicellular aggregation and persistence, is proposed to play an important role in the Salmonella transmission cycle. In this manuscript, we used luciferase reporters to examine the expression of csgD, which encodes the master biofilm regulator. We observed that the CsgD-regulated biofilm system responds differently to regulatory inputs once it is activated. Notably, the CsgD system became unresponsive to repression by Cpx and H-NS in high osmolarity conditions and less responsive to the addition of amino acids. Temperature-mediated regulation of csgD on agar was altered by intracellular levels of RpoS and cyclic-di-GMP. In contrast, the addition of glucose repressed CsgD biofilms seemingly independent of other signals. Understanding the fine-tuned regulation of csgD can help us to piece together how regulation occurs in natural environments, knowing that all Salmonella strains face strong selection pressures both within and outside their hosts. Ultimately, we can use this information to better control Salmonella and develop strategies to break the transmission cycle.
Keywords: CpxR; CsgD; Salmonella; biofilm; cellulose; curli.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kirk M.D., Pires S.M., Black R.E., Caipo M., Crump J.A., Devleesschauwer B., Döpfer D., Fazil A., Fischer-Walker C.L., Hald T., et al. Correction: World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PloS Med. 2015;12:e1001940. doi: 10.1371/journal.pmed.1001940. - DOI - PMC - PubMed
-
- Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M., The International Collaboration on Enteric Disease “Burden of Illness” Studies The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. - DOI - PubMed
-
- McClelland M., Sanderson K.E., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

