Quantitative Ultrastructural Morphometry and Gene Expression of mTOR-Related Mitochondriogenesis within Glioblastoma Cells
- PMID: 32604996
- PMCID: PMC7370179
- DOI: 10.3390/ijms21134570
Quantitative Ultrastructural Morphometry and Gene Expression of mTOR-Related Mitochondriogenesis within Glioblastoma Cells
Abstract
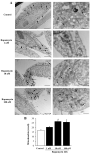
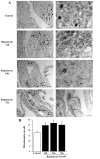
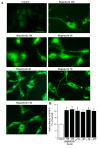
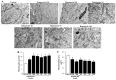
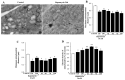
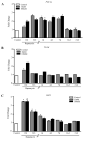
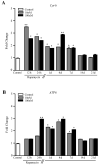
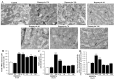
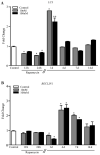
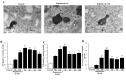
In glioblastoma (GBM) cells, an impairment of mitochondrial activity along with autophagy suppression occurs. Autophagy suppression in GBM promotes stemness, invasion, and poor prognosis. The autophagy deficit seems to be due, at least in part, to an abnormal up-regulation of the mammalian target of rapamycin (mTOR), which may be counteracted by pharmacological mTORC1 inhibition. Since autophagy activation is tightly bound to increased mitochondriogenesis, a defect in the synthesis of novel mitochondria is expected to occur in GBM cells. In an effort to measure a baseline deficit in mitochondria and promote mitochondriogenesis, the present study used two different GBM cell lines, both featuring mTOR hyperactivity. mTORC1 inhibition increases the expression of genes and proteins related to autophagy, mitophagy, and mitochondriogenesis. Autophagy activation was counted by RT-PCR of autophagy genes, LC3- immune-fluorescent puncta and immune-gold, as well as specific mitophagy-dependent BNIP3 stoichiometric increase in situ, within mitochondria. The activation of autophagy-related molecules and organelles after rapamycin exposure occurs concomitantly with progression of autophagosomes towards lysosomes. Remarkably, mitochondrial biogenesis and plasticity (increased mitochondrial number, integrity, and density as well as decreased mitochondrial area) was long- lasting for weeks following rapamycin withdrawal.
Keywords: NRF2; PGC1; autophagy; lysosomes; mitochondrial DNA; mitochondrial biogenesis; mitochondrial constitutive genes; mitophagy; rapamycin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









Similar articles
-
Rapamycin Ameliorates Defects in Mitochondrial Fission and Mitophagy in Glioblastoma Cells.Int J Mol Sci. 2021 May 20;22(10):5379. doi: 10.3390/ijms22105379. Int J Mol Sci. 2021. PMID: 34065350 Free PMC article.
-
Autophagy Activation Associates with Suppression of Prion Protein and Improved Mitochondrial Status in Glioblastoma Cells.Cells. 2023 Jan 4;12(2):221. doi: 10.3390/cells12020221. Cells. 2023. PMID: 36672156 Free PMC article.
-
Dcf1 induces glioblastoma cells apoptosis by blocking autophagy.Cancer Med. 2022 Jan;11(1):207-223. doi: 10.1002/cam4.4440. Epub 2021 Nov 19. Cancer Med. 2022. PMID: 34799992 Free PMC article.
-
Evidence that a mitochondrial death spiral underlies antagonistic pleiotropy.Aging Cell. 2017 Jun;16(3):435-443. doi: 10.1111/acel.12579. Epub 2017 Feb 9. Aging Cell. 2017. PMID: 28185435 Free PMC article. Review.
-
Mitochondrial clearance: mechanisms and roles in cellular fitness.FEBS Lett. 2021 Apr;595(8):1239-1263. doi: 10.1002/1873-3468.14060. Epub 2021 Mar 8. FEBS Lett. 2021. PMID: 33615465 Review.
Cited by
-
mTOR and SGLT-2 Inhibitors: Their Synergistic Effect on Age-Related Processes.Int J Mol Sci. 2024 Aug 8;25(16):8676. doi: 10.3390/ijms25168676. Int J Mol Sci. 2024. PMID: 39201363 Free PMC article. Review.
-
Merging the Multi-Target Effects of Phytochemicals in Neurodegeneration: From Oxidative Stress to Protein Aggregation and Inflammation.Antioxidants (Basel). 2020 Oct 20;9(10):1022. doi: 10.3390/antiox9101022. Antioxidants (Basel). 2020. PMID: 33092300 Free PMC article. Review.
-
Lactoferrin Protects against Methamphetamine Toxicity by Modulating Autophagy and Mitochondrial Status.Nutrients. 2021 Sep 25;13(10):3356. doi: 10.3390/nu13103356. Nutrients. 2021. PMID: 34684361 Free PMC article.
-
Alterations of Mitochondrial Structure in Methamphetamine Toxicity.Int J Mol Sci. 2022 Aug 10;23(16):8926. doi: 10.3390/ijms23168926. Int J Mol Sci. 2022. PMID: 36012188 Free PMC article.
-
In Silico Molecular Modeling to Identify the Role of Enzymes Involved in Mitochondriogenesis Upon Malvidin-3-Glucoside Effect.J Pharm Bioallied Sci. 2024 Apr;16(Suppl 2):S1295-S1298. doi: 10.4103/jpbs.jpbs_577_23. Epub 2024 Apr 16. J Pharm Bioallied Sci. 2024. PMID: 38882881 Free PMC article.
References
-
- Jiang H., White E.J., Conrad C., Gomez-Manzano C., Fueyo J. Autophagy pathways in glioblastoma. Methods Enzymol. 2009;453:273–286. - PubMed
-
- Huang X., Bai H.M., Chen L., Li B., Lu Y.C. Reduced expression of LC3B-II and Beclin 1 in glioblastoma multiforme indicates a down-regulated autophagic capacity that relates to the progression of astrocytic tumors. J. Clin. Neurosci. 2010;17:1515–1519. doi: 10.1016/j.jocn.2010.03.051. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

