Morphological and Mechanical Characterization of DNA SAMs Combining Nanolithography with AFM and Optical Methods
- PMID: 32605060
- PMCID: PMC7372444
- DOI: 10.3390/ma13132888
Morphological and Mechanical Characterization of DNA SAMs Combining Nanolithography with AFM and Optical Methods
Abstract
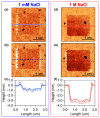
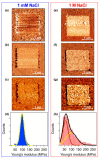
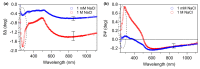
The morphological and mechanical properties of thiolated ssDNA films self-assembled at different ionic strength on flat gold surfaces have been investigated using Atomic Force Microscopy (AFM). AFM nanoshaving experiments, performed in hard tapping mode, allowed selectively removing molecules from micro-sized regions. To image the shaved areas, in addition to the soft contact mode, we explored the use of the Quantitative Imaging (QI) mode. QI is a less perturbative imaging mode that allows obtaining quantitative information on both sample topography and mechanical properties. AFM analysis showed that DNA SAMs assembled at high ionic strength are thicker and less deformable than films prepared at low ionic strength. In the case of thicker films, the difference between film and substrate Young's moduli could be assessed from the analysis of QI data. The AFM finding of thicker and denser films was confirmed by X-Ray Photoelectron Spectroscopy (XPS) and Spectroscopic Ellipsometry (SE) analysis. SE data allowed detecting the DNA UV absorption on dense monomolecular films. Moreover, feeding the SE analysis with the thickness data obtained by AFM, we could estimate the refractive index of dense DNA films.
Keywords: AFM; DNA; Spectroscopic Ellipsometry; ionic strength; molecular absorption; self-assembled monolayers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

