Multi-Laboratory Comparison of Next-Generation to Sanger-Based Sequencing for HIV-1 Drug Resistance Genotyping
- PMID: 32605062
- PMCID: PMC7411816
- DOI: 10.3390/v12070694
Multi-Laboratory Comparison of Next-Generation to Sanger-Based Sequencing for HIV-1 Drug Resistance Genotyping
Abstract
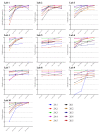
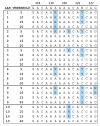
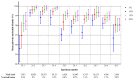
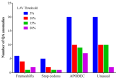
Next-generation sequencing (NGS) is increasingly used for HIV-1 drug resistance genotyping. NGS methods have the potential for a more sensitive detection of low-abundance variants (LAV) compared to standard Sanger sequencing (SS) methods. A standardized threshold for reporting LAV that generates data comparable to those derived from SS is needed to allow for the comparability of data from laboratories using NGS and SS. Ten HIV-1 specimens were tested in ten laboratories using Illumina MiSeq-based methods. The consensus sequences for each specimen using LAV thresholds of 5%, 10%, 15%, and 20% were compared to each other and to the consensus of the SS sequences (protease 4-99; reverse transcriptase 38-247). The concordance among laboratories' sequences at different thresholds was evaluated by pairwise sequence comparisons. NGS sequences generated using the 20% threshold were the most similar to the SS consensus (average 99.6% identity, range 96.1-100%), compared to 15% (99.4%, 88.5-100%), 10% (99.2%, 87.4-100%), or 5% (98.5%, 86.4-100%). The average sequence identity between laboratories using thresholds of 20%, 15%, 10%, and 5% was 99.1%, 98.7%, 98.3%, and 97.3%, respectively. Using the 20% threshold, we observed an excellent agreement between NGS and SS, but significant differences at lower thresholds. Understanding how variation in NGS methods influences sequence quality is essential for NGS-based HIV-1 drug resistance genotyping.
Keywords: HIV-1; NGS; drug resistance; genotyping.
Conflict of interest statement
N.P. is a consultant to the WHO HIVDR surveillance team and has performed contract work for Abbott Molecular, Aldatu Biosciences, Gilead Sciences, Roche Molecular Systems, Stanford University, and ThermoFisher Scientific. C.J.B. has received honoraria from Gilead Canada paid to his institution. The University of North Carolina is pursuing IP protection for Primer ID, and R.S. is listed as a co-inventor and has received nominal royalties.
Figures
References
-
- Li J.Z., Paredes R., Ribaudo H.J., Svarovskaia E.S., Metzner K.J., Kozal M.J., Hullsiek K.H., Balduin M., Jakobsen M.R., Geretti A.M., et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305:1327–1335. doi: 10.1001/jama.2011.375. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

