DNA Repair and Ovarian Carcinogenesis: Impact on Risk, Prognosis and Therapy Outcome
- PMID: 32605254
- PMCID: PMC7408288
- DOI: 10.3390/cancers12071713
DNA Repair and Ovarian Carcinogenesis: Impact on Risk, Prognosis and Therapy Outcome
Abstract
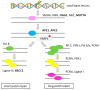
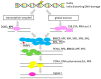
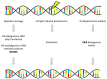
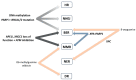
There is ample evidence for the essential involvement of DNA repair and DNA damage response in the onset of solid malignancies, including ovarian cancer. Indeed, highpenetrance germline mutations in DNA repair genes are important players in familial cancers: BRCA1, BRCA2 mutations or mismatch repair, and polymerase deficiency in colorectal, breast, and ovarian cancers. Recently, some molecular hallmarks (e.g., TP53, KRAS, BRAF, RAD51C/D or PTEN mutations) of ovarian carcinomas were identified. The manuscript overviews the role of DNA repair machinery in ovarian cancer, its risk, prognosis, and therapy outcome. We have attempted to expose molecular hallmarks of ovarian cancer with a focus on DNA repair system and scrutinized genetic, epigenetic, functional, and protein alterations in individual DNA repair pathways (homologous recombination, non-homologous end-joining, DNA mismatch repair, base- and nucleotide-excision repair, and direct repair). We suggest that lack of knowledge particularly in non-homologous end joining repair pathway and the interplay between DNA repair pathways needs to be confronted. The most important genes of the DNA repair system are emphasized and their targeting in ovarian cancer will deserve further attention. The function of those genes, as well as the functional status of the entire DNA repair pathways, should be investigated in detail in the near future.
Keywords: DNA repair; carcinogenesis; ovarian cancer; prognosis; therapy response.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

