Metabolic Signatures of Tumor Responses to Doxorubicin Elucidated by Metabolic Profiling in Ovo
- PMID: 32605263
- PMCID: PMC7408021
- DOI: 10.3390/metabo10070268
Metabolic Signatures of Tumor Responses to Doxorubicin Elucidated by Metabolic Profiling in Ovo
Abstract
Background: Dysregulated cancer metabolism is associated with acquired resistance to chemotherapeutic treatment and contributes to the activation of cancer survival mechanisms. However, which metabolic pathways are activated following treatment often remains elusive. The combination of chicken embryo tumor models (in ovo) with metabolomics phenotyping could offer a robust platform for drug testing. Here, we assess the potential of this approach in the treatment of an in ovo triple negative breast cancer with doxorubicin.
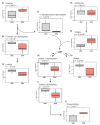
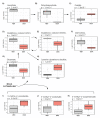
Methods: MB-MDA-231 cells were grafted in ovo. The resulting tumors were then treated with doxorubicin or dimethyl sulfoxide (DMSO) for six days. Tumors were collected and analyzed using a global untargeted metabolomics and comprehensive lipidomics.
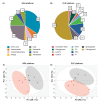
Results: We observed a significant suppression of tumor growth in the doxorubicin treated group. The metabolic profiles of doxorubicin and DMSO-treated tumors were clearly separated in a principle component analysis. Inhibition of glycolysis, nucleotide synthesis, and glycerophospholipid metabolism appear to be triggered by doxorubicin treatment, which could explain the observed suppressed tumor growth. In addition, metabolic cancer survival mechanisms could be supported by an acceleration of antioxidative pathways.
Conclusions: Metabolomics in combination with in ovo tumor models provide a robust platform for drug testing to reveal tumor specific treatment targets such as the antioxidative tumor capacity.
Keywords: cancer survival mechanism; chicken chorioallantoic membrane (CAM) system in ovo model; doxorubicin treatment; lipidomics; metabolomics; triple negative breast cancer.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
-
- Malorni L., Shetty P.B., De Angelis C., Hilsenbeck S., Rimawi M.F., Elledge R., Osborne C.K., De Placido S., Arpino G. Clinical and biologic features of triple−negative breast cancers in a large cohort of patients with long−term follow−up. Breast Cancer Res. Treat. 2012;136:795–804. doi: 10.1007/s10549-012-2315-y. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

