Genome assembly of the basket willow, Salix viminalis, reveals earliest stages of sex chromosome expansion
- PMID: 32605573
- PMCID: PMC7329446
- DOI: 10.1186/s12915-020-00808-1
Genome assembly of the basket willow, Salix viminalis, reveals earliest stages of sex chromosome expansion
Abstract
Background: Sex chromosomes have evolved independently multiple times in eukaryotes and are therefore considered a prime example of convergent genome evolution. Sex chromosomes are known to emerge after recombination is halted between a homologous pair of chromosomes, and this leads to a range of non-adaptive modifications causing gradual degeneration and gene loss on the sex-limited chromosome. However, the proximal causes of recombination suppression and the pace at which degeneration subsequently occurs remain unclear.
Results: Here, we use long- and short-read single-molecule sequencing approaches to assemble and annotate a draft genome of the basket willow, Salix viminalis, a species with a female heterogametic system at the earliest stages of sex chromosome emergence. Our single-molecule approach allowed us to phase the emerging Z and W haplotypes in a female, and we detected very low levels of Z/W single-nucleotide divergence in the non-recombining region. Linked-read sequencing of the same female and an additional male (ZZ) revealed the presence of two evolutionary strata supported by both divergence between the Z and W haplotypes and by haplotype phylogenetic trees. Gene order is still largely conserved between the Z and W homologs, although the W-linked region contains genes involved in cytokinin signaling regulation that are not syntenic with the Z homolog. Furthermore, we find no support across multiple lines of evidence for inversions, which have long been assumed to halt recombination between the sex chromosomes.
Conclusions: Our data suggest that selection against recombination is a more gradual process at the earliest stages of sex chromosome formation than would be expected from an inversion and may result instead from the accumulation of transposable elements. Our results present a cohesive understanding of the earliest genomic consequences of recombination suppression as well as valuable insights into the initial stages of sex chromosome formation and regulation of sex differentiation.
Keywords: Recombination suppression; Salix; Sex chromosomes; W-chromosome; Willow.
Conflict of interest statement
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare that they have no competing interests.
Figures
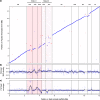
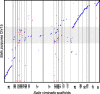
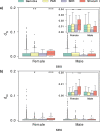


References
-
- Beukeboom LW, Perrin N. The evolution of sex determination. Oxford: Oxford University Press; 2014. p. 222.
Publication types
MeSH terms
Grants and funding
- 680951/ERC_/European Research Council/International
- 2016-20031/Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas)/International
- 30599-5/Swedish Energy Agency/International
- 260233/ERC_/European Research Council/International
- 150/Canada Research Chairs/International
LinkOut - more resources
Full Text Sources

