Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties
- PMID: 32606306
- PMCID: PMC7327033
- DOI: 10.1038/s41467-020-17047-1
Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties
Abstract
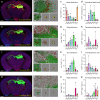
Recombinant adeno-associated viruses (rAAVs) are currently considered the safest and most reliable gene delivery vehicles for human gene therapy. Three serotype capsids, AAV1, AAV2, and AAV9, have been approved for commercial use in patients, but they may not be suitable for all therapeutic contexts. Here, we describe a novel capsid identified in a human clinical sample by high-throughput, long-read sequencing. The capsid, which we have named AAVv66, shares high sequence similarity with AAV2. We demonstrate that compared to AAV2, AAVv66 exhibits enhanced production yields, virion stability, and CNS transduction. Unique structural properties of AAVv66 visualized by cryo-EM at 2.5-Å resolution, suggest that critical residues at the three-fold protrusion and at the interface of the five-fold axis of symmetry likely contribute to the beneficial characteristics of AAVv66. Our findings underscore the potential of AAVv66 as a gene therapy vector.
Conflict of interest statement
G.G. is a scientific co-founder of Voyager Therapeutics and holds equity in the company. G.G. is an inventor on patents with potential royalties licensed to Voyager Therapeutics and other biopharmaceutical companies. G.G. and D.J.G. are scientific co-founders of Aspa Therapeutics Inc., and hold equity in the company. G.G. and D.J.G. are inventors on patents with potential royalties licensed to Aspa Therapeutics Inc., and other biopharmaceutical companies. Remaining authors declare no competing interests.
Figures





References
-
- Muzyczka N, Berns K. Parvoviridae: the viruses and their replication. Fields Virol. 2001;2:2327–2359.
-
- Grieger JC, Johnson JS, Gurda-Whitaker B, Agbandje-McKenna M, Samulski RJ. Surface-exposed adeno-associated virus Vp1-NLS capsid fusion protein rescues infectivity of noninfectious wild-type Vp2/Vp3 and Vp3-only capsids but not that of fivefold pore mutant virions. J. Virol. 2007;81:7833–7843. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

