Evaluation of population stratification adjustment using genome-wide or exonic variants
- PMID: 32608112
- PMCID: PMC7722041
- DOI: 10.1002/gepi.22332
Evaluation of population stratification adjustment using genome-wide or exonic variants
Abstract
Population stratification may cause an inflated type-I error and spurious association when assessing the association between genetic variations with an outcome. Many genetic association studies are now using exonic variants, which captures only 1% of the genome, however, population stratification adjustments have not been evaluated in the context of exonic variants. We compare the performance of two established approaches: principal components analysis (PCA) and mixed-effects models and assess the utility of genome-wide (GW) and exonic variants, by simulation and using a data set from the Framingham Heart Study. Our results illustrate that although the PCs and genetic relationship matrices computed by GW and exonic markers are different, the type-I error rate of association tests for common variants with additive effect appear to be properly controlled in the presence of population stratification. In addition, by considering single nucleotide variants (SNVs) that have different levels of confounding by population stratification, we also compare the power across multiple association approaches to account for population stratification such as PC-based corrections and mixed-effects models. We find that while these two methods achieve a similar power for SNVs that have a low or medium level of confounding by population stratification, mixed-effects model can reach a higher power for SNVs highly confounded by population stratification.
Keywords: GWAS; PCA; mixed-effects model; population stratification.
© 2020 Wiley Periodicals LLC.
Figures
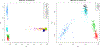
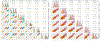
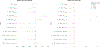

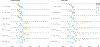

Similar articles
-
Improving the power of GWAS and avoiding confounding from population stratification with PC-Select.Genetics. 2014 Jul;197(3):1045-9. doi: 10.1534/genetics.114.164285. Epub 2014 Apr 29. Genetics. 2014. PMID: 24788602 Free PMC article.
-
Accounting for population stratification in DNA methylation studies.Genet Epidemiol. 2014 Apr;38(3):231-41. doi: 10.1002/gepi.21789. Epub 2014 Jan 29. Genet Epidemiol. 2014. PMID: 24478250 Free PMC article.
-
Evaluation of methods for adjusting population stratification in genome-wide association studies: Standard versus categorical principal component analysis.Ann Hum Genet. 2019 Nov;83(6):454-464. doi: 10.1111/ahg.12339. Epub 2019 Jul 19. Ann Hum Genet. 2019. PMID: 31322288
-
New approaches to population stratification in genome-wide association studies.Nat Rev Genet. 2010 Jul;11(7):459-63. doi: 10.1038/nrg2813. Nat Rev Genet. 2010. PMID: 20548291 Free PMC article. Review.
-
Software engineering the mixed model for genome-wide association studies on large samples.Brief Bioinform. 2009 Nov;10(6):664-75. doi: 10.1093/bib/bbp050. Brief Bioinform. 2009. PMID: 19933212 Review.
Cited by
-
Polygenic Risk for Insomnia in Adolescents of Diverse Ancestry.Front Genet. 2021 May 10;12:654717. doi: 10.3389/fgene.2021.654717. eCollection 2021. Front Genet. 2021. PMID: 34040634 Free PMC article.
-
Population stratification correction using Bayesian shrinkage priors for genetic association studies.Ann Hum Genet. 2023 Nov;87(6):302-315. doi: 10.1111/ahg.12527. Epub 2023 Sep 28. Ann Hum Genet. 2023. PMID: 37771252 Free PMC article.
References
-
- Belkadi Aziz, Pedergnana Vincent, Cobat Aurélie, Itan Yuval, Vincent Quentin B., Abhyankar Avinash, . . . Abel Laurent. (2016). Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proceedings of the National Academy of Sciences of the United States of America, 113(24), 6713–6718. doi:10.1073/pnas.1606460113 - DOI - PMC - PubMed
-
- Chen W, Chen H, Wang C, Conomos M, Stilp A, Li Z, . . . Lin X. (2016). Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. The American Journal of Human Genetics, 98(4), 653–666. doi:10.1016/j.ajhg.2016.02.012 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

