CDKD-dependent activation of CDKA;1 controls microtubule dynamics and cytokinesis during meiosis
- PMID: 32609301
- PMCID: PMC7401817
- DOI: 10.1083/jcb.201907016
CDKD-dependent activation of CDKA;1 controls microtubule dynamics and cytokinesis during meiosis
Abstract
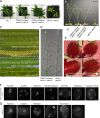
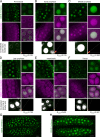
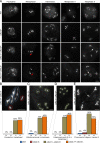
Precise control of cytoskeleton dynamics and its tight coordination with chromosomal events are key to cell division. This is exemplified by formation of the spindle and execution of cytokinesis after nuclear division. Here, we reveal that the central cell cycle regulator CYCLIN DEPENDENT KINASE A;1 (CDKA;1), the Arabidopsis homologue of Cdk1 and Cdk2, partially in conjunction with CYCLIN B3;1 (CYCB3;1), is a key regulator of the microtubule cytoskeleton in meiosis. For full CDKA;1 activity, the function of three redundantly acting CDK-activating kinases (CAKs), CDKD;1, CDKD;2, and CDKD;3, is necessary. Progressive loss of these genes in combination with a weak loss-of-function mutant in CDKA;1 allowed a fine-grained dissection of the requirement of cell-cycle kinase activity for meiosis. Notably, a moderate reduction of CDKA;1 activity converts the simultaneous cytokinesis in Arabidopsis, i.e., one cytokinesis separating all four meiotic products concurrently into two successive cytokineses with cell wall formation after the first and second meiotic division, as found in many monocotyledonous species.
© 2020 Sofroni et al.
Figures











References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

