Tyramide signal amplification mass spectrometry (TSA-MS) ratio identifies nuclear speckle proteins
- PMID: 32609799
- PMCID: PMC7480118
- DOI: 10.1083/jcb.201910207
Tyramide signal amplification mass spectrometry (TSA-MS) ratio identifies nuclear speckle proteins
Abstract
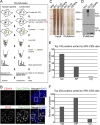
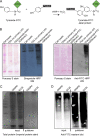
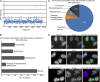
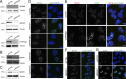
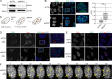
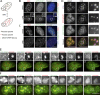
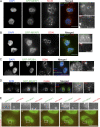
We present a simple ratio method to infer protein composition within cellular structures using proximity labeling approaches but compensating for the diffusion of free radicals. We used tyramide signal amplification (TSA) and label-free mass spectrometry (MS) to compare proteins in nuclear speckles versus centromeres. Our "TSA-MS ratio" approach successfully identified known nuclear speckle proteins. For example, 96% and 67% of proteins in the top 30 and 100 sorted proteins, respectively, are known nuclear speckle proteins, including proteins that we validated here as enriched in nuclear speckles. We show that MFAP1, among the top 20 in our list, forms droplets under certain circumstances and that MFAP1 expression levels modulate the size, stability, and dynamics of nuclear speckles. Localization of MFAP1 and its binding partner, PRPF38A, in droplet-like nuclear bodies precedes formation of nuclear speckles during telophase. Our results update older proteomic studies of nuclear speckles and should provide a useful reference dataset to guide future experimental dissection of nuclear speckle structure and function.
© 2020 Dopie et al.
Figures


References
-
- Baldin V., Militello M., Thomas Y., Doucet C., Fic W., Boireau S., Jariel-Encontre I., Piechaczyk M., Bertrand E., Tazi J., et al. . 2008. A novel role for PA28γ-proteasome in nuclear speckle organization and SR protein trafficking. Mol. Biol. Cell. 19:1706–1716. 10.1091/mbc.e07-07-0637 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

