Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis
- PMID: 32610096
- PMCID: PMC7311890
- DOI: 10.1016/j.cmet.2020.06.016
Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis
Abstract
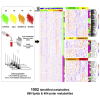
The coronavirus disease 2019 (COVID-19) pandemic presents an unprecedented threat to global public health. Herein, we utilized a combination of targeted and untargeted tandem mass spectrometry to analyze the plasma lipidome and metabolome in mild, moderate, and severe COVID-19 patients and healthy controls. A panel of 10 plasma metabolites effectively distinguished COVID-19 patients from healthy controls (AUC = 0.975). Plasma lipidome of COVID-19 resembled that of monosialodihexosyl ganglioside (GM3)-enriched exosomes, with enhanced levels of sphingomyelins (SMs) and GM3s, and reduced diacylglycerols (DAGs). Systems evaluation of metabolic dysregulation in COVID-19 was performed using multiscale embedded differential correlation network analyses. Using exosomes isolated from the same cohort, we demonstrated that exosomes of COVID-19 patients with elevating disease severity were increasingly enriched in GM3s. Our work suggests that GM3-enriched exosomes may partake in pathological processes related to COVID-19 pathogenesis and presents the largest repository on the plasma lipidome and metabolome distinct to COVID-19.
Keywords: COVID-19; biomarker; bis(monoacylglyero)phosphates; exosomes; lipidomics; metabolomics; monosialodihexosyl gangliosides; phosphatidylserines.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests S.M.L., G.H.C., and B.L. are employees of LipidALL Technologies.
Figures




References
-
- Baez J.M., Barbour S.E., Cohen D.E. Phosphatidylcholine transfer protein promotes apolipoprotein A-I-mediated lipid efflux in Chinese hamster ovary cells. J. Biol. Chem. 2002;277:6198–6206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

