A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome
- PMID: 32612235
- PMCID: PMC7610377
- DOI: 10.1038/s41586-020-2444-0
A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome
Abstract
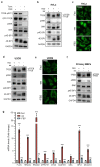
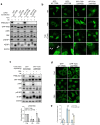
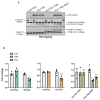
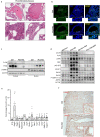
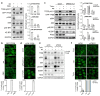
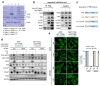
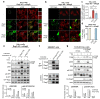
The mechanistic target of rapamycin complex 1 (mTORC1) is a key metabolic hub that controls the cellular response to environmental cues by exerting its kinase activity on multiple substrates1-3. However, whether mTORC1 responds to diverse stimuli by differentially phosphorylating specific substrates is poorly understood. Here we show that transcription factor EB (TFEB), a master regulator of lysosomal biogenesis and autophagy4,5, is phosphorylated by mTORC1 via a substrate-specific mechanism that is mediated by Rag GTPases. Owing to this mechanism, the phosphorylation of TFEB-unlike other substrates of mTORC1, such as S6K and 4E-BP1- is strictly dependent on the amino-acid-mediated activation of RagC and RagD GTPases, but is insensitive to RHEB activity induced by growth factors. This mechanism has a crucial role in Birt-Hogg-Dubé syndrome, a disorder that is caused by mutations in the RagC and RagD activator folliculin (FLCN) and is characterized by benign skin tumours, lung and kidney cysts and renal cell carcinoma6,7. We found that constitutive activation of TFEB is the main driver of the kidney abnormalities and mTORC1 hyperactivity in a mouse model of Birt-Hogg-Dubé syndrome. Accordingly, depletion of TFEB in kidneys of these mice fully rescued the disease phenotype and associated lethality, and normalized mTORC1 activity. Our findings identify a mechanism that enables differential phosphorylation of mTORC1 substrates, the dysregulation of which leads to kidney cysts and cancer.
Conflict of interest statement
A.B. is Co-Founder of CASMA Therapeutics, Inc, Cambridge, MA 02139
Figures







Comment in
-
Differential mTORC1 pathways in BHD.Nat Rev Urol. 2020 Sep;17(9):485. doi: 10.1038/s41585-020-0364-2. Nat Rev Urol. 2020. PMID: 32733039 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

