Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion
- PMID: 32612238
- PMCID: PMC7116900
- DOI: 10.1038/s41586-020-2454-y
Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion
Abstract
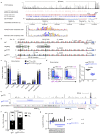
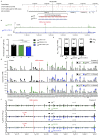
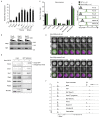
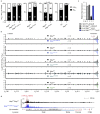
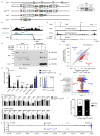
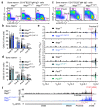
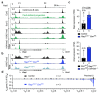
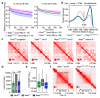
Nuclear processes, such as V(D)J recombination, are orchestrated by the three-dimensional organization of chromosomes at multiple levels, including compartments1 and topologically associated domains (TADs)2,3 consisting of chromatin loops4. TADs are formed by chromatin-loop extrusion5-7, which depends on the loop-extrusion function of the ring-shaped cohesin complex8-12. Conversely, the cohesin-release factor Wapl13,14 restricts loop extension10,15. The generation of a diverse antibody repertoire, providing humoral immunity to pathogens, requires the participation of all V genes in V(D)J recombination16, which depends on contraction of the 2.8-Mb-long immunoglobulin heavy chain (Igh) locus by Pax517,18. However, how Pax5 controls Igh contraction in pro-B cells remains unknown. Here we demonstrate that locus contraction is caused by loop extrusion across the entire Igh locus. Notably, the expression of Wapl is repressed by Pax5 specifically in pro-B and pre-B cells, facilitating extended loop extrusion by increasing the residence time of cohesin on chromatin. Pax5 mediates the transcriptional repression of Wapl through a single Pax5-binding site by recruiting the polycomb repressive complex 2 to induce bivalent chromatin at the Wapl promoter. Reduced Wapl expression causes global alterations in the chromosome architecture, indicating that the potential to recombine all V genes entails structural changes of the entire genome in pro-B cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures







References
-
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

