Deep learning methods in protein structure prediction
- PMID: 32612753
- PMCID: PMC7305407
- DOI: 10.1016/j.csbj.2019.12.011
Deep learning methods in protein structure prediction
Abstract
Protein Structure Prediction is a central topic in Structural Bioinformatics. Since the '60s statistical methods, followed by increasingly complex Machine Learning and recently Deep Learning methods, have been employed to predict protein structural information at various levels of detail. In this review, we briefly introduce the problem of protein structure prediction and essential elements of Deep Learning (such as Convolutional Neural Networks, Recurrent Neural Networks and basic feed-forward Neural Networks they are founded on), after which we discuss the evolution of predictive methods for one-dimensional and two-dimensional Protein Structure Annotations, from the simple statistical methods of the early days, to the computationally intensive highly-sophisticated Deep Learning algorithms of the last decade. In the process, we review the growth of the databases these algorithms are based on, and how this has impacted our ability to leverage knowledge about evolution and co-evolution to achieve improved predictions. We conclude this review outlining the current role of Deep Learning techniques within the wider pipelines to predict protein structures and trying to anticipate what challenges and opportunities may arise next.
Keywords: Deep learning; Machine learning; Protein structure prediction.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
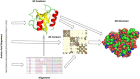
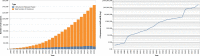
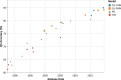
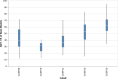
References
-
- Kendrew J.C., Dickerson R.E., Strandberg B.E., Hart R.G., Davies D.R., Phillips D.C., Shore V.C. Structure of myoglobin: a three-dimensional Fourier synthesis at 2 A. resolution. Nature. 1960;185:422–427. - PubMed
-
- Perutz M.F., Rossmann M.G., Cullis A.F., Muirhead H., Will G., North A.C. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960;185:416–422. - PubMed
-
- Siegel J.B., Zanghellini A., Lovick H.M., Kiss G., Lambert A.R., St.Clair J.L., Gallaher J.L., Hilvert D., Gelb M.H., Stoddard B.L., Houk K.N., Michael F.E., Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular diels-alder reaction. Science. 2010;329:309–313. - PMC - PubMed
-
- Kuhlman B., Dantas G., Ireton G.C., Varani G., Stoddard B.L., Baker D. Design of a Novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
