A hands-on introduction to querying evolutionary relationships across multiple data sources using SPARQL
- PMID: 32612807
- PMCID: PMC7324951
- DOI: 10.12688/f1000research.21027.2
A hands-on introduction to querying evolutionary relationships across multiple data sources using SPARQL
Abstract
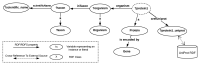
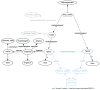
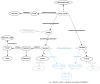
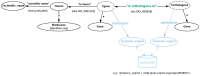
The increasing use of Semantic Web technologies in the life sciences, in particular the use of the Resource Description Framework (RDF) and the RDF query language SPARQL, opens the path for novel integrative analyses, combining information from multiple data sources. However, analyzing evolutionary data in RDF is not trivial, due to the steep learning curve required to understand both the data models adopted by different RDF data sources, as well as the equivalent SPARQL constructs required to benefit from this data - in particular, recursive property paths. In this article, we provide a hands-on introduction to querying evolutionary data across several data sources that publish orthology information in RDF, namely: The Orthologous MAtrix (OMA), the European Bioinformatics Institute (EBI) RDF platform, the Database of Orthologous Groups (OrthoDB) and the Microbial Genome Database (MBGD). We present four protocols in increasing order of complexity. In these protocols, we demonstrate through SPARQL queries how to retrieve pairwise orthologs, homologous groups, and hierarchical orthologous groups. Finally, we show how orthology information in different data sources can be compared, through the use of federated SPARQL queries.
Keywords: Comparative Genomics; Orthology; Resource Description Framework (RDF); SPARQL; Sequence Homology.
Copyright: © 2020 Sima AC et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

References
-
- de Farias TM, Chiba H, Fernández-Breis JT: Leveraging logical rules for efficacious representation of large orthology datasets. s.l., Proceedings of the 10th International Semantic Web Applications and Tools for Healthcare and Life Sciences (SWAT4HCLS) Conference2017. Reference Source
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

