Identification of novel genetic factors underlying the host-pathogen interaction between barley (Hordeum vulgare L.) and powdery mildew (Blumeria graminis f. sp. hordei)
- PMID: 32614894
- PMCID: PMC7332009
- DOI: 10.1371/journal.pone.0235565
Identification of novel genetic factors underlying the host-pathogen interaction between barley (Hordeum vulgare L.) and powdery mildew (Blumeria graminis f. sp. hordei)
Abstract
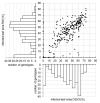
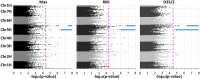
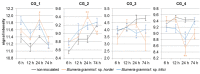
Powdery mildew is an important foliar disease of barley (Hordeum vulgare L.) caused by the biotrophic fungus Blumeria graminis f. sp. hordei (Bgh). The understanding of the resistance mechanism is essential for future resistance breeding. In particular, the identification of race-nonspecific resistance genes is important because of their regarded durability and broad-spectrum activity. We assessed the severity of powdery mildew infection on detached seedling leaves of 267 barley accessions using two poly-virulent isolates and performed a genome-wide association study exploiting 201 of these accessions. Two-hundred and fourteen markers, located on six barley chromosomes are associated with potential race-nonspecific Bgh resistance or susceptibility. Initial steps for the functional validation of four promising candidates were performed based on phenotype and transcription data. Specific candidate alleles were analyzed via transient gene silencing as well as transient overexpression. Microarray data of the four selected candidates indicate differential regulation of the transcription in response to Bgh infection. Based on our results, all four candidate genes seem to be involved in the responses to powdery mildew attack. In particular, the transient overexpression of specific alleles of two candidate genes, a potential arabinogalactan protein and the barley homolog of Arabidopsis thaliana's Light-Response Bric-a-Brac/-Tramtrack/-Broad Complex/-POxvirus and Zinc finger (AtLRB1) or AtLRB2, were top candidates of novel powdery mildew susceptibility genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Specific Resistance of Barley to Powdery Mildew, Its Use and Beyond. A Concise Critical Review.Genes (Basel). 2020 Aug 21;11(9):971. doi: 10.3390/genes11090971. Genes (Basel). 2020. PMID: 32825722 Free PMC article. Review.
-
Small RNA discovery in the interaction between barley and the powdery mildew pathogen.BMC Genomics. 2019 Jul 25;20(1):610. doi: 10.1186/s12864-019-5947-z. BMC Genomics. 2019. PMID: 31345162 Free PMC article.
-
Interaction of a Blumeria graminis f. sp. hordei effector candidate with a barley ARF-GAP suggests that host vesicle trafficking is a fungal pathogenicity target.Mol Plant Pathol. 2014 Aug;15(6):535-49. doi: 10.1111/mpp.12110. Epub 2014 Mar 3. Mol Plant Pathol. 2014. PMID: 24304971 Free PMC article.
-
Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen.Proc Natl Acad Sci U S A. 2016 Oct 18;113(42):E6486-E6495. doi: 10.1073/pnas.1612947113. Epub 2016 Oct 4. Proc Natl Acad Sci U S A. 2016. PMID: 27702901 Free PMC article.
-
Magical mystery tour: MLO proteins in plant immunity and beyond.New Phytol. 2014 Oct;204(2):273-81. doi: 10.1111/nph.12889. New Phytol. 2014. PMID: 25453131 Review.
Cited by
-
Inference of Host-Pathogen Interaction Matrices from Genome-Wide Polymorphism Data.Mol Biol Evol. 2024 Sep 4;41(9):msae176. doi: 10.1093/molbev/msae176. Mol Biol Evol. 2024. PMID: 39172738 Free PMC article.
-
Specific Resistance of Barley to Powdery Mildew, Its Use and Beyond. A Concise Critical Review.Genes (Basel). 2020 Aug 21;11(9):971. doi: 10.3390/genes11090971. Genes (Basel). 2020. PMID: 32825722 Free PMC article. Review.
-
Genome-wide association mapping highlights candidate genes and immune genotypes for net blotch and powdery mildew resistance in barley.Comput Struct Biotechnol J. 2023 Oct 10;21:4923-4932. doi: 10.1016/j.csbj.2023.10.014. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37867969 Free PMC article.
-
Insights into the Genetic Architecture and Genomic Prediction of Powdery Mildew Resistance in Flax (Linum usitatissimum L.).Int J Mol Sci. 2022 Apr 29;23(9):4960. doi: 10.3390/ijms23094960. Int J Mol Sci. 2022. PMID: 35563347 Free PMC article.
-
Advancing fungal phylogenetics: integrating modern sequencing, dark taxa discovery, and machine learning.Arch Microbiol. 2025 Jul 11;207(9):192. doi: 10.1007/s00203-025-04392-2. Arch Microbiol. 2025. PMID: 40643763 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

