CD81 Controls Beige Fat Progenitor Cell Growth and Energy Balance via FAK Signaling
- PMID: 32615086
- PMCID: PMC7415677
- DOI: 10.1016/j.cell.2020.06.021
CD81 Controls Beige Fat Progenitor Cell Growth and Energy Balance via FAK Signaling
Abstract
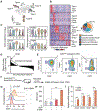
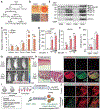
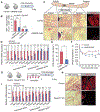
Adipose tissues dynamically remodel their cellular composition in response to external cues by stimulating beige adipocyte biogenesis; however, the developmental origin and pathways regulating this process remain insufficiently understood owing to adipose tissue heterogeneity. Here, we employed single-cell RNA-seq and identified a unique subset of adipocyte progenitor cells (APCs) that possessed the cell-intrinsic plasticity to give rise to beige fat. This beige APC population is proliferative and marked by cell-surface proteins, including PDGFRα, Sca1, and CD81. Notably, CD81 is not only a beige APC marker but also required for de novo beige fat biogenesis following cold exposure. CD81 forms a complex with αV/β1 and αV/β5 integrins and mediates the activation of integrin-FAK signaling in response to irisin. Importantly, CD81 loss causes diet-induced obesity, insulin resistance, and adipose tissue inflammation. These results suggest that CD81 functions as a key sensor of external inputs and controls beige APC proliferation and whole-body energy homeostasis.
Keywords: adipocyte progenitors; adipogenesis; beige fat; brown fat; diabetes; metabolic adaptation; metabolic disease; metabolism; obesity; tissue remodeling.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



Comment in
-
Finding a Needle in a Haystack: Identification of a Beige Fat Progenitor.Cell. 2020 Aug 6;182(3):537-539. doi: 10.1016/j.cell.2020.07.014. Cell. 2020. PMID: 32763184
References
-
- Berry DC, Jiang Y, Arpke RW, Close EL, Uchida A, Reading D, Berglund ED, Kyba M, and Graff JM (2017). Cellular Aging Contributes to Failure of Cold-Induced Beige Adipocyte Formation in Old Mice and Humans. Cell metabolism 25, 481. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK063720/DK/NIDDK NIH HHS/United States
- M01 RR001271/RR/NCRR NIH HHS/United States
- P30 DK098722/DK/NIDDK NIH HHS/United States
- P30 DK020541/DK/NIDDK NIH HHS/United States
- T32 HL007374/HL/NHLBI NIH HHS/United States
- R01 DK112268/DK/NIDDK NIH HHS/United States
- K99 DK110426/DK/NIDDK NIH HHS/United States
- UL1 TR000004/TR/NCATS NIH HHS/United States
- R00 DK110426/DK/NIDDK NIH HHS/United States
- R01 DK097441/DK/NIDDK NIH HHS/United States
- R01 DK125281/DK/NIDDK NIH HHS/United States
- P30 DK026687/DK/NIDDK NIH HHS/United States
- R01 DK110098/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

