Methane, arsenic, selenium and the origins of the DMSO reductase family
- PMID: 32616801
- PMCID: PMC7331816
- DOI: 10.1038/s41598-020-67892-9
Methane, arsenic, selenium and the origins of the DMSO reductase family
Abstract
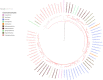
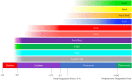
Mononuclear molybdoenzymes of the dimethyl sulfoxide reductase (DMSOR) family catalyze a number of reactions essential to the carbon, nitrogen, sulfur, arsenic, and selenium biogeochemical cycles. These enzymes are also ancient, with many lineages likely predating the divergence of the last universal common ancestor into the Bacteria and Archaea domains. We have constructed rooted phylogenies for over 1,550 representatives of the DMSOR family using maximum likelihood methods to investigate the evolution of the arsenic biogeochemical cycle. The phylogenetic analysis provides compelling evidence that formylmethanofuran dehydrogenase B subunits, which catalyze the reduction of CO2 to formate during hydrogenotrophic methanogenesis, constitutes the most ancient lineage. Our analysis also provides robust support for selenocysteine as the ancestral ligand for the Mo/W atom. Finally, we demonstrate that anaerobic arsenite oxidase and respiratory arsenate reductase catalytic subunits represent a more ancient lineage of DMSORs compared to aerobic arsenite oxidase catalytic subunits, which evolved from the assimilatory nitrate reductase lineage. This provides substantial support for an active arsenic biogeochemical cycle on the anoxic Archean Earth. Our work emphasizes that the use of chalcophilic elements as substrates as well as the Mo/W ligand in DMSORs has indelibly shaped the diversification of these enzymes through deep time.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Schoepp-Cothenet B, et al. On the universal core of bioenergetics. Biochim. Biophys. Acta. 2013;1827:79–93. - PubMed
-
- Grimaldi S, Schoepp-Cothenet B, Ceccaldi P, Guigliarelli B, Magalon A. The prokaryotic Mo/W-bisPGD enzymes family: A catalytic workhorse in bioenergetic. Biochim. Biophys. Acta. 2013;1827:1048–1085. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

