Elongating maize root: zone-specific combinations of polysaccharides from type I and type II primary cell walls
- PMID: 32616810
- PMCID: PMC7331734
- DOI: 10.1038/s41598-020-67782-0
Elongating maize root: zone-specific combinations of polysaccharides from type I and type II primary cell walls
Abstract
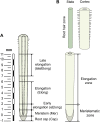
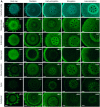
The dynamics of cell wall polysaccharides may modulate the cell wall mechanics and thus control the expansion growth of plant cells. The unique composition of type II primary cell wall characteristic of grasses suggests that they employ specific mechanisms for cell enlargement. We characterized the transcriptomes in five zones along maize root, clustered the expression of genes for numerous glycosyltransferases and performed extensive immunohistochemical analysis to relate the changes in cell wall polysaccharides to critical stages of cell development in Poaceae. Specific patterns of cell wall formation differentiate the initiation, realization and cessation of elongation growth. Cell walls of meristem and early elongation zone represent a mixture of type I and type II specific polysaccharides. Xyloglucans and homogalacturonans are synthesized there actively together with mixed-linkage glucans and glucuronoarabinoxylans. Rhamnogalacturonans-I with the side-chains of branched 1,4-galactan and arabinan persisted in cell walls throughout the development. Thus, the machinery to generate the type I primary cell wall constituents is completely established and operates. The expression of glycosyltransferases responsible for mixed-linkage glucan and glucuronoarabinoxylan synthesis peaks at active or late elongation. These findings widen the number of jigsaw pieces which should be put together to solve the puzzle of grass cell growth.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

