BRICseq Bridges Brain-wide Interregional Connectivity to Neural Activity and Gene Expression in Single Animals
- PMID: 32619423
- PMCID: PMC7771207
- DOI: 10.1016/j.cell.2020.05.029
BRICseq Bridges Brain-wide Interregional Connectivity to Neural Activity and Gene Expression in Single Animals
Erratum in
-
BRICseq Bridges Brain-wide Interregional Connectivity to Neural Activity and Gene Expression in Single Animals.Cell. 2020 Dec 23;183(7):2040. doi: 10.1016/j.cell.2020.12.009. Cell. 2020. PMID: 33357401 Free PMC article. No abstract available.
Abstract
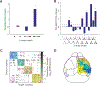
Comprehensive analysis of neuronal networks requires brain-wide measurement of connectivity, activity, and gene expression. Although high-throughput methods are available for mapping brain-wide activity and transcriptomes, comparable methods for mapping region-to-region connectivity remain slow and expensive because they require averaging across hundreds of brains. Here we describe BRICseq (brain-wide individual animal connectome sequencing), which leverages DNA barcoding and sequencing to map connectivity from single individuals in a few weeks and at low cost. Applying BRICseq to the mouse neocortex, we find that region-to-region connectivity provides a simple bridge relating transcriptome to activity: the spatial expression patterns of a few genes predict region-to-region connectivity, and connectivity predicts activity correlations. We also exploited BRICseq to map the mutant BTBR mouse brain, which lacks a corpus callosum, and recapitulated its known connectopathies. BRICseq allows individual laboratories to compare how age, sex, environment, genetics, and species affect neuronal wiring and to integrate these with functional activity and gene expression.
Keywords: BRICseq; MAPseq; connectome; high-throughput sequencing; mesoscale.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests A.M.Z. is a founder of Cajal Neuroscience and a member of its scientific advisory board.
Figures
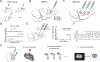
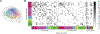
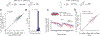



Comment in
-
Connectomics in high throughput.Nat Methods. 2020 Sep;17(9):873. doi: 10.1038/s41592-020-0948-z. Nat Methods. 2020. PMID: 32873982 No abstract available.
References
-
- Banerjee A, Phelps SM, and Long MA (2019). Singing mice. Curr. Biol 29, R190–R191. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

