Naive Human Embryonic Stem Cells Can Give Rise to Cells with a Trophoblast-like Transcriptome and Methylome
- PMID: 32619492
- PMCID: PMC7363941
- DOI: 10.1016/j.stemcr.2020.06.003
Naive Human Embryonic Stem Cells Can Give Rise to Cells with a Trophoblast-like Transcriptome and Methylome
Abstract
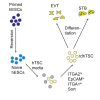
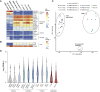
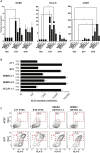
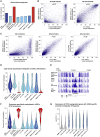
Human embryonic stem cells (hESCs) readily differentiate to somatic or germ lineages but have impaired ability to form extra-embryonic lineages such as placenta or yolk sac. Here, we demonstrate that naive hESCs can be converted into cells that exhibit the cellular and molecular phenotypes of human trophoblast stem cells (hTSCs) derived from human placenta or blastocyst. The resulting "transdifferentiated" hTSCs show reactivation of core placental genes, acquisition of a placenta-like methylome, and the ability to differentiate to extravillous trophoblasts and syncytiotrophoblasts. Modest differences are observed between transdifferentiated and placental hTSCs, most notably in the expression of certain imprinted loci. These results suggest that naive hESCs can differentiate to extra-embryonic lineage and demonstrate a new way of modeling human trophoblast specification and placental methylome establishment.
Keywords: DNA methylation; amnion; development; differentiation; embryonic stem cells; epigenetics; placenta; pluripotency; trophoblast.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

