HEBE, a novel positive regulator of senescence in Solanum lycopersicum
- PMID: 32620827
- PMCID: PMC7335192
- DOI: 10.1038/s41598-020-67937-z
HEBE, a novel positive regulator of senescence in Solanum lycopersicum
Abstract
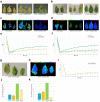
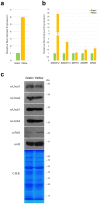
Leaf senescence and plant aging are traits of great interest for breeders. Senescing cells undergo important physiological and biochemical changes, while cellular structures such as chloroplasts are degraded with dramatic metabolic consequences for the whole plant. The possibility of prolonging the photosynthetic ability of leaves could positively impact the plant's life span with benefits for biomass production and metabolite accumulation; plants with these characteristics display a stay-green phenotype. A group of plant transcription factors known as NAC play a pivotal role in controlling senescence: here we describe the involvement of the tomato NAC transcription factor Solyc12g036480, which transcript is present in leaves and floral buds. Since its silencing delays leaf senescence and prevents plants from ageing, we renamed Solyc12g0364 HḖBĒ, for the Greek goddess of youth. In this manuscript we describe how HEB downregulation negatively affects the progression of senescence, resulting in changes in transcription of senescence-promoting genes, as well as the activity of enzymes involved in chlorophyll degradation, thereby explaining the stay-green phenotype.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Buchanan-Wollaston V. Senescence processes in plants. Annual plant review, volume 26. Ann. Bot. 2008;101:197.
-
- Gregersen PL, Holm PB, Krupinska K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol. 2008;10(Suppl 1):37–49. - PubMed

