Engineering monocyte/macrophage-specific glucocerebrosidase expression in human hematopoietic stem cells using genome editing
- PMID: 32620863
- PMCID: PMC7335164
- DOI: 10.1038/s41467-020-17148-x
Engineering monocyte/macrophage-specific glucocerebrosidase expression in human hematopoietic stem cells using genome editing
Erratum in
-
Author Correction: Engineering monocyte/macrophage-specific glucocerebrosidase expression in human hematopoietic stem cells using genome editing.Nat Commun. 2020 Aug 20;11(1):4231. doi: 10.1038/s41467-020-18044-0. Nat Commun. 2020. PMID: 32820153 Free PMC article.
Abstract
Gaucher disease is a lysosomal storage disorder caused by insufficient glucocerebrosidase activity. Its hallmark manifestations are attributed to infiltration and inflammation by macrophages. Current therapies for Gaucher disease include life-long intravenous administration of recombinant glucocerebrosidase and orally-available glucosylceramide synthase inhibitors. An alternative approach is to engineer the patient's own hematopoietic system to restore glucocerebrosidase expression, thereby replacing the affected cells, and constituting a potential one-time therapy for this disease. Here, we report an efficient CRISPR/Cas9-based approach that targets glucocerebrosidase expression cassettes with a monocyte/macrophage-specific element to the CCR5 safe-harbor locus in human hematopoietic stem and progenitor cells. The targeted cells generate glucocerebrosidase-expressing macrophages and maintain long-term repopulation and multi-lineage differentiation potential with serial transplantation. The combination of a safe-harbor and a lineage-specific promoter establishes a universal correction strategy and circumvents potential toxicity of ectopic glucocerebrosidase in the stem cells. Furthermore, it constitutes an adaptable platform for other lysosomal enzyme deficiencies.
Conflict of interest statement
M.H.P. declares that he is a consultant and has equity interest in CRISPR Tx and Allogene Tx, and he states that neither company has had input or opinions on the subject matter described in this manuscript. The other authors declare no competing interests.
Figures


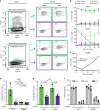
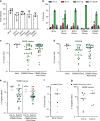


References
-
- Pastores, G. M. & Hughes, D. A. in GeneReviews((R)). (eds Adam, M. P. et al.) (University of Washington, Seattle University of Washington, Seattle, 1993). GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved., Seattle (WA)).
-
- Charrow J, et al. The Gaucher registry: demographics and disease characteristics of 1698 patients with Gaucher disease. Arch. Intern. Med. 2000;160:2835–2843. - PubMed
-
- Ferraz MJ, et al. Gaucher disease and Fabry disease: new markers and insights in pathophysiology for two distinct glycosphingolipidoses. Biochim. Biophys. Acta. 2014;1841:811–825. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

