Fold-stratified cross-validation for unbiased and privacy-preserving federated learning
- PMID: 32620945
- PMCID: PMC7647321
- DOI: 10.1093/jamia/ocaa096
Fold-stratified cross-validation for unbiased and privacy-preserving federated learning
Abstract
Objective: We introduce fold-stratified cross-validation, a validation methodology that is compatible with privacy-preserving federated learning and that prevents data leakage caused by duplicates of electronic health records (EHRs).
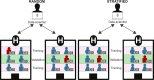
Materials and methods: Fold-stratified cross-validation complements cross-validation with an initial stratification of EHRs in folds containing patients with similar characteristics, thus ensuring that duplicates of a record are jointly present either in training or in validation folds. Monte Carlo simulations are performed to investigate the properties of fold-stratified cross-validation in the case of a model data analysis using both synthetic data and MIMIC-III (Medical Information Mart for Intensive Care-III) medical records.
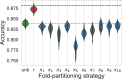
Results: In situations in which duplicated EHRs could induce overoptimistic estimations of accuracy, applying fold-stratified cross-validation prevented this bias, while not requiring full deduplication. However, a pessimistic bias might appear if the covariate used for the stratification was strongly associated with the outcome.
Discussion: Although fold-stratified cross-validation presents low computational overhead, to be efficient it requires the preliminary identification of a covariate that is both shared by duplicated records and weakly associated with the outcome. When available, the hash of a personal identifier or a patient's date of birth provides such a covariate. On the contrary, pseudonymization interferes with fold-stratified cross-validation, as it may break the equality of the stratifying covariate among duplicates.
Conclusion: Fold-stratified cross-validation is an easy-to-implement methodology that prevents data leakage when a model is trained on distributed EHRs that contain duplicates, while preserving privacy.
Keywords: data leakage; duplicated electronic health records; federated learning; privacy; validation.
© The Author(s) 2020. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Komorowski M, Celi LA, Badawi O, et al. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med 2018; 24 (11): 1716–20. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

