Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies
- PMID: 32623480
- PMCID: PMC7334636
- DOI: 10.1007/s00018-020-03580-1
Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies
Abstract
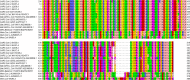
The recent severe acute respiratory syndrome, known as Coronavirus Disease 2019 (COVID-19) has spread so much rapidly and severely to induce World Health Organization (WHO) to declare a state of emergency over the new coronavirus SARS-CoV-2 pandemic. While several countries have chosen the almost complete lock-down for slowing down SARS-CoV-2 spread, the scientific community is called to respond to the devastating outbreak by identifying new tools for diagnosis and treatment of the dangerous COVID-19. With this aim, we performed an in silico comparative modeling analysis, which allows gaining new insights into the main conformational changes occurring in the SARS-CoV-2 spike protein, at the level of the receptor-binding domain (RBD), along interactions with human cells angiotensin-converting enzyme 2 (ACE2) receptor, that favor human cell invasion. Furthermore, our analysis provides (1) an ideal pipeline to identify already characterized antibodies that might target SARS-CoV-2 spike RBD, aiming to prevent interactions with the human ACE2, and (2) instructions for building new possible neutralizing antibodies, according to chemical/physical space restraints and complementary determining regions (CDR) mutagenesis of the identified existing antibodies. The proposed antibodies show in silico high affinity for SARS-CoV-2 spike RBD and can be used as reference antibodies also for building new high-affinity antibodies against present and future coronaviruses able to invade human cells through interactions of their spike proteins with the human ACE2. More in general, our analysis provides indications for the set-up of the right biological molecular context for investigating spike RBD-ACE2 interactions for the development of new vaccines, diagnostic kits, and other treatments based on the targeting of SARS-CoV-2 spike protein.
Keywords: ACE2 and ACE inhibitors; COVID-19; Comparative modeling; Coronavirus; Fold recognition tools; Neutralizing antibodies; Receptor binding domain; SARS-CoV-2; Spike; Spike post-fusion conformation; n-CoV19.
Figures

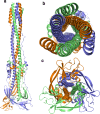



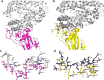
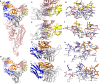
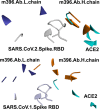

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

