Glioblastoma Myeloid-Derived Suppressor Cell Subsets Express Differential Macrophage Migration Inhibitory Factor Receptor Profiles That Can Be Targeted to Reduce Immune Suppression
- PMID: 32625208
- PMCID: PMC7315581
- DOI: 10.3389/fimmu.2020.01191
Glioblastoma Myeloid-Derived Suppressor Cell Subsets Express Differential Macrophage Migration Inhibitory Factor Receptor Profiles That Can Be Targeted to Reduce Immune Suppression
Abstract
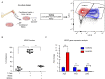
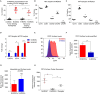
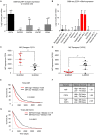
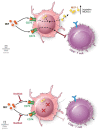
The application of tumor immunotherapy to glioblastoma (GBM) is limited by an unprecedented degree of immune suppression due to factors that include high numbers of immune suppressive myeloid cells, the blood brain barrier, and T cell sequestration to the bone marrow. We previously identified an increase in immune suppressive myeloid-derived suppressor cells (MDSCs) in GBM patients, which correlated with poor prognosis and was dependent on macrophage migration inhibitory factor (MIF). Here we examine the MIF signaling axis in detail in murine MDSC models, GBM-educated MDSCs and human GBM. We found that the monocytic subset of MDSCs (M-MDSCs) expressed high levels of the MIF cognate receptor CD74 and was localized in the tumor microenvironment. In contrast, granulocytic MDSCs (G-MDSCs) expressed high levels of the MIF non-cognate receptor CXCR2 and showed minimal accumulation in the tumor microenvironment. Furthermore, targeting M-MDSCs with Ibudilast, a brain penetrant MIF-CD74 interaction inhibitor, reduced MDSC function and enhanced CD8 T cell activity in the tumor microenvironment. These findings demonstrate the MDSC subsets differentially express MIF receptors and may be leveraged for specific MDSC targeting.
Keywords: MDSC; MIF–macrophage migration inhibitory factor; glioma; immunesuppresion; immunetherapy.
Copyright © 2020 Alban, Bayik, Otvos, Rabljenovic, Leng, Jia-Shiun, Roversi, Lauko, Momin, Mohammadi, Peereboom, Ahluwalia, Matsuda, Yun, Bucala, Vogelbaum and Lathia.
Figures


References
-
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. (2009) 10:459–66. 10.1016/S1470-20450970025-7 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

