Genetic resistance to transmissible spongiform encephalopathies (TSE) in goats
- PMID: 32625625
- PMCID: PMC7010077
- DOI: 10.2903/j.efsa.2017.4962
Genetic resistance to transmissible spongiform encephalopathies (TSE) in goats
Abstract
Breeding programmes to promote resistance to classical scrapie, similar to those for sheep in existing transmissible spongiform encephalopathies (TSE) regulations, have not been established in goats. The European Commission requested a scientific opinion from EFSA on the current knowledge of genetic resistance to TSE in goats. An evaluation tool, which considers both the weight of evidence and strength of resistance to classical scrapie of alleles in the goat PRNP gene, was developed and applied to nine selected alleles of interest. Using the tool, the quality and certainty of the field and experimental data are considered robust enough to conclude that the K222, D146 and S146 alleles both confer genetic resistance against classical scrapie strains known to occur naturally in the EU goat population, with which they have been challenged both experimentally and under field conditions. The weight of evidence for K222 is greater than that currently available for the D146 and S146 alleles and for the ARR allele in sheep in 2001. Breeding for resistance can be an effective tool for controlling classical scrapie in goats and it could be an option available to member states, both at herd and population levels. There is insufficient evidence to assess the impact of K222, D146 and S146 alleles on susceptibility to atypical scrapie and bovine spongiform encephalopathy (BSE), or on health and production traits. These alleles are heterogeneously distributed across the EU Member States and goat breeds, but often at low frequencies (< 10%). Given these low frequencies, high selection pressure may have an adverse effect on genetic diversity so any breeding for resistance programmes should be developed at Member States, rather than EU level and their impact monitored, with particular attention to the potential for any negative impact in rare or small population breeds.
Keywords: TSE; classical; genetics; goats; resistance; scrapie.
© 2017 European Food Safety Authority. EFSA Journal published by John Wiley and Sons Ltd on behalf of European Food Safety Authority.
Figures
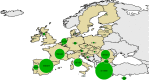


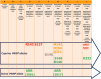
Green:
MS that reported bothCS andAS . Blue:MS that reported onlyCS . Yellow:MS that reported onlyAS . White:MS that have not reported caprine scrapie.
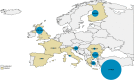
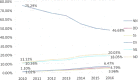
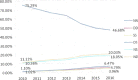
Number of cases/10,000 rapid tests standardised by stream, i.e.,
SHC vs.NSHC during the period 2002–2015. The proportion of tests carried out in all the 28MS s in theNSHC vsSHC in goats has been used to define the baseline population for the direct standardisation. The sizes of the blue dots are proportional to the prevalence.

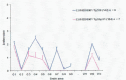
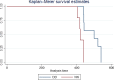
Crosses (+) indicate the annual stream‐adjusted prevalence (cases per 10,000 rapid tests). Lines show the linear trend (black line) and the bounds of the 95%
CI (grey lines).
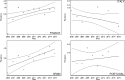
Crosses (+) indicate the annual stream‐adjusted prevalence (cases per 10,000 rapid tests). Lines show the linear trend (black line) and the bounds of the 95%
CI (grey lines).

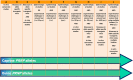


*The selection regime is based on allele frequency and population size (lower right corner), whereby under mild selection heterozygous and homozygous bucks are used indiscriminately, under moderate selection homozygous rams are used preferably but supplemented with heterozygous bucks, and under strong selection homozygous rams are used exclusively. Limits for classifying populations as large, medium or small have not been determined in goat breeds, but were < 750 for small breeds and > 3,750 for large breeds in sheep. Scheme designed by Jack J. Windig.

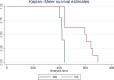
Only clinically positive mice contributed to the lesion profiles. Ten mice of each mouse line were challenged. Numbers in brackets indicate the average
IP of the mice which contributed to the profile. n = number of mice which contributed to the profile.



References
-
- Acín C, Martin‐Burriel I, Monleon E, Lyahyai J, Pitarch JL, Serrano C, Monzon M, Zaragoza P and Badiola JJ, 2013. Prion protein gene variability in Spanish goats. Inference through susceptibility to classical scrapie strains and pathogenic distribution of peripheral PrP(sc.). PLoS ONE, 8, e61118. - PMC - PubMed
-
- Acutis PL, Bossers A, Priem J, Riina MV, Peletto S, Mazza M, Casalone C, Forloni G, Ru G and Caramelli M, 2006. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks. Journal of General Virology, 87, 1029–1033. - PubMed
-
- Acutis PL, Colussi S, Santagada G, Laurenza C, Maniaci MG, Riina MV, Peletto S, Goldmann W, Bossers A, Caramelli M and Cristoferi I, 2008. Genetic variability of the PRNP gene in goat breeds from Northern and Southern Italy. Journal of Applied Microbiology, 104, 1782–1789. - PubMed
-
- Acutis PL, Martucci F, D'Angelo A, Peletto S, Colussi S, Maurella C, Porcario C, Iulini B, Mazza M, Dell'atti L, Zuccon F, Corona C, Martinelli N, Casalone C, Caramelli M and Lombardi G, 2012. Resistance to classical scrapie in experimentally challenged goats carrying mutation K222 of the prion protein gene. Veterinary Research, 43, 8. - PMC - PubMed
-
- Agrimi U, Ru G, Cardone F, Pocchiari M and Caramelli M, 1999. Epidemic of transmissible spongiform encephalopathy in sheep and goats in Italy. Lancet, 353, 560–561. - PubMed
LinkOut - more resources
Full Text Sources
