Identification of the Mutational Landscape of Gynecological Malignancies
- PMID: 32626534
- PMCID: PMC7330690
- DOI: 10.7150/jca.46174
Identification of the Mutational Landscape of Gynecological Malignancies
Abstract
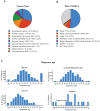
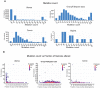
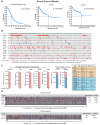
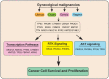
Background: Cancer is a complex disease that arises from the accumulation of multiple genetic and non-genetic changes. Advances in sequencing technologies have allowed unbiased and global analysis of patient-derived tumor samples and the discovery of genetic and transcriptional changes in key genes and oncogenic pathways. That in turn has facilitated a better understanding of the underlying causes of cancer initiation and progression, resulting in new therapeutic targets. Methods: In our study, we have analyzed the mutational landscape of gynecological malignancies using datasets from The Cancer Genome Atlas (TCGA). We have also analyzed Oncomine datasets to establish the impact of their alteration on disease recurrence and survival of patients. Results: In this study, we analyzed a series of different gynecological malignancies for commonly occurring genetic and non-genetic alterations. These studies show that white women have higher incidence of gynecological malignancies. Furthermore, our study identified 16 genes that are altered at a frequency >10% among all of the gynecological malignancies and tumor suppressor TP53 is the most altered gene in these malignancies (>50% of the cases). The top 16 genes fall into the categories of either tumor suppressor or oncogenes and a subset of these genes are associated with poor prognosis, some affecting recurrence and survival of ovarian cancer patients. Conclusion: In sum, our study identified 16 major genes that are broadly mutated in a large majority of gynecological malignancies and in some cases predict survival and recurrence in patients with gynecological malignancies. We predict that the functional studies will determine their relative role in the initiation and progression of gynecological malignancies and also establish if some of them represents drug targets for anti-cancer therapy.
Keywords: Oncomine; TCGA; biomarker; gynecological malignancies; therapeutic.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

