A prospective study on rapid exome sequencing as a diagnostic test for multiple congenital anomalies on fetal ultrasound
- PMID: 32627857
- PMCID: PMC7540374
- DOI: 10.1002/pd.5781
A prospective study on rapid exome sequencing as a diagnostic test for multiple congenital anomalies on fetal ultrasound
Abstract
Objective: Conventional genetic tests (quantitative fluorescent-PCR [QF-PCR] and single nucleotide polymorphism-array) only diagnose ~40% of fetuses showing ultrasound abnormalities. Rapid exome sequencing (rES) may improve this diagnostic yield, but includes challenges such as uncertainties in fetal phenotyping, variant interpretation, incidental unsolicited findings, and rapid turnaround times. In this study, we implemented rES in prenatal care to increase diagnostic yield.
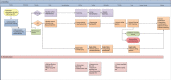
Methods: We prospectively studied 55 fetuses. Inclusion criteria were: (a) two or more independent major fetal anomalies, (b) hydrops fetalis or bilateral renal cysts alone, or (c) one major fetal anomaly and a first-degree relative with the same anomaly. In addition to conventional genetic tests, we performed trio rES analysis using a custom virtual gene panel of ~3850 Online Mendelian Inheritance in Man (OMIM) genes.
Results: We established a genetic rES-based diagnosis in 8 out of 23 fetuses (35%) without QF-PCR or array abnormalities. Diagnoses included MIRAGE (SAMD9), Zellweger (PEX1), Walker-Warburg (POMGNT1), Noonan (PTNP11), Kabuki (KMT2D), and CHARGE (CHD7) syndrome and two cases of Osteogenesis Imperfecta type 2 (COL1A1). In six cases, rES diagnosis aided perinatal management. The median turnaround time was 14 (range 8-20) days.
Conclusion: Implementing rES as a routine test in the prenatal setting is challenging but technically feasible, with a promising diagnostic yield and significant clinical relevance.
© 2020 The Authors. Prenatal Diagnosis published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Eurocat update: short report congenital anomalies Northern Netherlands 1981–2017 (update); 2019. www.eurocat.umcg.nl
-
- Fleurke‐Rozema JH, Bakker MK, Snijders RJM, Bilardo CM. Uptake of the 20‐week scan and the detection rate of selected sonomarkers and structural congenital anomalies (Chapter 8 thesis) In: Fleurke‐Rozema JH, ed. Impact of the 20‐week scan. Groningen, The Netherlands: Rijksuniversiteit Groningen; 2017. https://www.rug.nl/research/portal/files/46538014/Chapter_8.pdf.
-
- The International Society for Prenatal Diagnosis , The Society for Maternal and Fetal Medicine , The Perinatal Quality Foundation . Joint Position Statement from the International Society for Prenatal Diagnosis (ISPD), the Society for Maternal Fetal Medicine (SMFM), and the Perinatal Quality Foundation (PQF) on the use of genome‐wide sequencing for fetal diagnosis. Prenatal Diagn. 2018;38(1):6‐9. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

