Facets of individual-specific health signatures determined from longitudinal plasma proteome profiling
- PMID: 32629387
- PMCID: PMC7334812
- DOI: 10.1016/j.ebiom.2020.102854
Facets of individual-specific health signatures determined from longitudinal plasma proteome profiling
Abstract
Background: Precision medicine approaches aim to tackle diseases on an individual level through molecular profiling. Despite the growing knowledge about diseases and the reported diversity of molecular phenotypes, the descriptions of human health on an individual level have been far less elaborate.
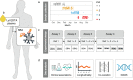
Methods: To provide insights into the longitudinal protein signatures of well-being, we profiled blood plasma collected over one year from 101 clinically healthy individuals using multiplexed antibody assays. After applying an antibody validation scheme, we utilized > 700 protein profiles for in-depth analyses of the individuals' short-term health trajectories.
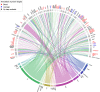
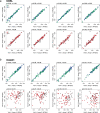
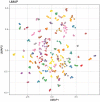
Findings: We found signatures of circulating proteomes to be highly individual-specific. Considering technical and longitudinal variability, we observed that 49% of the protein profiles were stable over one year. We also identified eight networks of proteins in which 11-242 proteins covaried over time. For each participant, there were unique protein profiles of which some could be explained by associations to genetic variants.
Interpretation: This observational and non-interventional study identifyed noticeable diversity among clinically healthy subjects, and facets of individual-specific signatures emerged by monitoring the variability of the circulating proteomes over time. To enable more personal hence precise assessments of health states, longitudinal profiling of circulating proteomes can provide a valuable component for precision medicine approaches.
Funding: This work was supported by the Erling Persson Foundation, the Swedish Heart and Lung Foundation, the Knut and Alice Wallenberg Foundation, Science for Life Laboratory, and the Swedish Research Council.
Keywords: Affinity proteomics; Longitudinal profiling; Plasma proteomics; Precision medicine; pQTLs.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Figures



References
-
- Hu JX, Thomas CE, Brunak S. Network biology concepts in complex disease comorbidities. Nat Rev Genet. 2016;17(10):615–629. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

