Cannabinoids and Cannabinoid Receptors: The Story so Far
- PMID: 32629422
- PMCID: PMC7339067
- DOI: 10.1016/j.isci.2020.101301
Cannabinoids and Cannabinoid Receptors: The Story so Far
Abstract
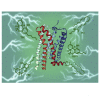
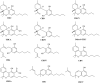
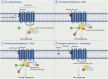
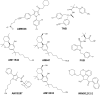
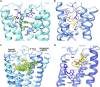
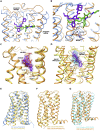
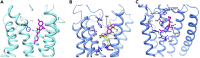
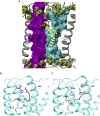
Like most modern molecular biology and natural product chemistry, understanding cannabinoid pharmacology centers around molecular interactions, in this case, between the cannabinoids and their putative targets, the G-protein coupled receptors (GPCRs) cannabinoid receptor 1 (CB1) and cannabinoid receptor 2 (CB2). Understanding the complex structure and interplay between the partners in this molecular dance is required to understand the mechanism of action of synthetic, endogenous, and phytochemical cannabinoids. This review, with 91 references, surveys our understanding of the structural biology of the cannabinoids and their target receptors including both a critical comparison of the extant crystal structures and the computationally derived homology models, as well as an in-depth discussion about the binding modes of the major cannabinoids. The aim is to assist in situating structural biochemists, synthetic chemists, and molecular biologists who are new to the field of cannabis research.
Keywords: Medical Substance; Molecular Biology; Structural Biology; Supramolecular Chemistry.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Baker D., Pryce G., Giovannoni G., Thompson A.J. The therapeutic potential of cannabis. Lancet Neurol. 2003;2:291–298. - PubMed
-
- Bouaboula M., Poinot-Chazel C., Marchand J., Canat X., Bourrié B., Rinaldi-Carmona M., Calandra B., Le Fur G., Casellas P. Signaling pathway associated with stimulation of CB2 peripheral cannabinoid receptor. Eur. J. Biochem. 1996;237:704–711. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

