A Comparative Study of Engineered Dermal Templates for Skin Wound Repair in a Mouse Model
- PMID: 32630398
- PMCID: PMC7350005
- DOI: 10.3390/ijms21124508
A Comparative Study of Engineered Dermal Templates for Skin Wound Repair in a Mouse Model
Abstract
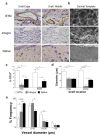
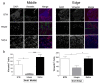
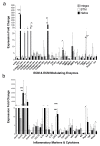
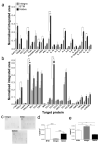
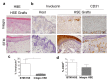
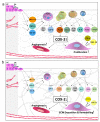
Engineered dermal templates have revolutionised the repair and reconstruction of skin defects. Their interaction with the wound microenvironment and linked molecular mediators of wound repair is still not clear. This study investigated the wound bed and acellular "off the shelf" dermal template interaction in a mouse model. Full-thickness wounds in nude mice were grafted with allogenic skin, and either collagen-based or fully synthetic dermal templates. Changes in the wound bed showed significantly higher vascularisation and fibroblast infiltration in synthetic grafts when compared to collagen-based grafts (P ≤ 0.05). Greater tissue growth was associated with higher prostaglandin-endoperoxide synthase 2 (Ptgs2) RNA and cyclooxygenase-2 (COX-2) protein levels in fully synthetic grafts. Collagen-based grafts had higher levels of collagen III and matrix metallopeptidase 2. To compare the capacity to form a double layer skin substitute, both templates were seeded with human fibroblasts and keratinocytes (so-called human skin equivalent or HSE). Mice were grafted with HSEs to test permanent wound closure with no further treatment required. We found the synthetic dermal template to have a significantly greater capacity to support human epidermal cells. In conclusion, the synthetic template showed advantages over the collagen-based template in a short-term mouse model of wound repair.
Keywords: COX-2; Integra®; NovoSorb® BTM; dermal templates; graft; human skin equivalent; inflammation; wound repair.
Conflict of interest statement
Authors have no conflict of interest to declare.
Figures
References
-
- Rygaard J. Skin grafts in nude mice. 1. Allografts in nude mice of three genetic backgrounds (BALB-c, C3H, C57-BL) Acta Pathol. Microbiol. Scand. A. 1974;82:80–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

