SARS-CoV-2: Repurposed Drugs and Novel Therapeutic Approaches-Insights into Chemical Structure-Biological Activity and Toxicological Screening
- PMID: 32630746
- PMCID: PMC7409030
- DOI: 10.3390/jcm9072084
SARS-CoV-2: Repurposed Drugs and Novel Therapeutic Approaches-Insights into Chemical Structure-Biological Activity and Toxicological Screening
Abstract
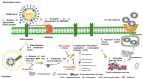
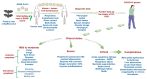
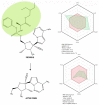
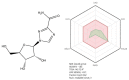
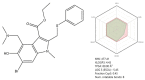
SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) pandemic represents the primary public health concern nowadays, and great efforts are made worldwide for efficient management of this crisis. Considerable scientific progress was recorded regarding SARS-CoV-2 infection in terms of genomic structure, diagnostic tools, viral transmission, mechanism of viral infection, symptomatology, clinical impact, and complications, but these data evolve constantly. Up to date, neither an effective vaccine nor SARS-CoV-2 specific antiviral agents have been approved, but significant advances were enlisted in this direction by investigating repurposed approved drugs (ongoing clinical trials) or developing innovative antiviral drugs (preclinical and clinical studies). This review presents a thorough analysis of repurposed drug admitted for compassionate use from a chemical structure-biological activity perspective highlighting the ADME (absorption, distribution, metabolism, and excretion) properties and the toxicophore groups linked to potential adverse effects. A detailed pharmacological description of the novel potential anti-COVID-19 therapeutics was also included. In addition, a comprehensible overview of SARS-CoV-2 infection in terms of general description and structure, mechanism of viral infection, and clinical impact was portrayed.
Keywords: SARS-CoV-2; chloroquine; convalescent plasma; hydroxychloroquine; lopinavir/ritonavir; remdesivir.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


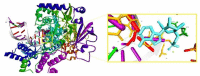

References
-
- Lai C.C., Liu Y.H., Wang C.Y., Wang Y.H., Hsueh S.C., Yen M.Y., Ko W.C., Hsueh P.R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.02.012. - DOI - PMC - PubMed
-
- World Health Organization Home Page—WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. [(accessed on 25 April 2020)]; Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-re....
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

