Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing
- PMID: 32630835
- PMCID: PMC7407193
- DOI: 10.3390/cells9071608
Latest Developed Strategies to Minimize the Off-Target Effects in CRISPR-Cas-Mediated Genome Editing
Abstract
Gene editing that makes target gene modification in the genome by deletion or addition has revolutionized the era of biomedicine. Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 emerged as a substantial tool due to its simplicity in use, less cost and extraordinary efficiency than the conventional gene-editing tools, including zinc finger nucleases (ZFNs) and Transcription activator-like effector nucleases (TALENs). However, potential off-target activities are crucial shortcomings in the CRISPR system. Numerous types of approaches have been developed to reduce off-target effects. Here, we review several latest approaches to reduce the off-target effects, including biased or unbiased off-target detection, cytosine or adenine base editors, prime editing, dCas9, Cas9 paired nickase, ribonucleoprotein (RNP) delivery and truncated gRNAs. This review article provides extensive information to cautiously interpret off-target effects to assist the basic and clinical applications in biomedicine.
Keywords: CRISPR/Cas9; base editors; gene targeting; homology dependent repair; non-homologous end joining repair pathway; targeting specificity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
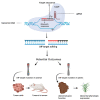
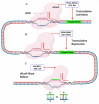
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

