PDXGEM: patient-derived tumor xenograft-based gene expression model for predicting clinical response to anticancer therapy in cancer patients
- PMID: 32631229
- PMCID: PMC7336455
- DOI: 10.1186/s12859-020-03633-z
PDXGEM: patient-derived tumor xenograft-based gene expression model for predicting clinical response to anticancer therapy in cancer patients
Abstract
Background: Cancer is a highly heterogeneous disease with varying responses to anti-cancer drugs. Although several attempts have been made to predict the anti-cancer therapeutic responses, there remains a great need to develop highly accurate prediction models of response to the anti-cancer drugs for clinical applications toward a personalized medicine. Patient derived xenografts (PDXs) are preclinical cancer models in which the tissue or cells from a patient's tumor are implanted into an immunodeficient or humanized mouse. In the present study, we develop a bioinformatics analysis pipeline to build a predictive gene expression model (GEM) for cancer patients' drug responses based on gene expression and drug activity data from PDX models.
Results: Drug sensitivity biomarkers were identified by performing an association analysis between gene expression levels and post-treatment tumor volume changes in PDX models. We built a drug response prediction model (called PDXGEM) in a random-forest algorithm by using a subset of the drug sensitvity biomarkers with concordant co-expression patterns between the PDXs and pretreatment cancer patient tumors. We applied the PDXGEM to several cytotoxic chemotherapies as well as targeted therapy agents that are used to treat breast cancer, pancreatic cancer, colorectal cancer, or non-small cell lung cancer. Significantly accurate predictions of PDXGEM for pathological response or survival outcomes were observed in extensive independent validations on multiple cancer patient datasets obtained from retrospective observational studies and prospective clinical trials.
Conclusion: Our results demonstrated the strong potential of using molecular profiles and drug activity data of PDX tumors in developing a clinically translatable predictive cancer biomarkers for cancer patients. The PDXGEM web application is publicly available at http://pdxgem.moffitt.org .
Keywords: Chemotherapy; Drug response prediction; Gene expression; PDX; Patient-derived xenograft model; Predictive cancer biomarker; Targeted therapy.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

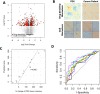

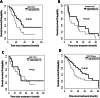

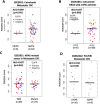

References
-
- Rupp T, Zuckerman D. Quality of life, overall survival, and costs of Cancer drugs approved based on surrogate endpoints. JAMA Intern Med. 2017;177:276–277. - PubMed
-
- Zou J, Wang E. Cancer biomarker discovery for precision medicine: new progresses. Curr Med Chem. 2018;26:7655–7671. - PubMed
-
- Boyd MR. In: The NCI in vitro anticancer drug discovery screen. in anticancer drug development guide: preclinical screening, clinical trials, and approval. Teicher BA, editor. Totowa: Humana Press; 1997. pp. 23–42.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

