Structural basis for the stabilization of amyloidogenic immunoglobulin light chains by hydantoins
- PMID: 32631553
- PMCID: PMC7402200
- DOI: 10.1016/j.bmcl.2020.127356
Structural basis for the stabilization of amyloidogenic immunoglobulin light chains by hydantoins
Abstract
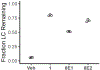
Misfolding and aggregation of immunoglobulin light chains (LCs) leads to the degeneration of post-mitotic tissue in the disease immunoglobulin LC amyloidosis (AL). We previously reported the discovery of small molecule kinetic stabilizers of the native dimeric structure of full-length LCs, which slow or stop the LC aggregation cascade at the outset. A predominant structural category of kinetic stabilizers emerging from the high-throughput screen are coumarins substituted at the 7-position, which bind at the interface between the two variable domains of the light chain dimer. Here, we report the binding mode of another, more polar, LC kinetic stabilizer chemotype, 3,5-substituted hydantoins. Computational docking, solution nuclear magnetic resonance experiments, and x-ray crystallography show that the aromatic substructure emerging from the hydantoin 3-position occupies the same LC binding site as the coumarin ring. Notably, the hydantoin ring extends beyond the binding site mapped out by the coumarin hits. The hydantoin ring makes hydrogen bonds with both LC monomers simultaneously. The alkyl substructure at the hydantoin 5-position partially occupies a novel binding pocket proximal to the pocket occupied by the coumarin substructure. Overall, the hydantoin structural data suggest that a larger area of the LC variable-domain-variable-domain dimer interface is amenable to small molecule binding than previously demonstrated, which should facilitate development of more potent full-length LC kinetic stabilizers.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
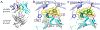
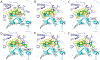



References
-
- Kelly JW, Clinical Trial Support for the Hypothesis that the Process of Protein Aggregation Causes Neurodegeneration. Sci Transl Med, 2020.
-
- Hammarstrom P, et al., Prevention of transthyret in amyloid disease by changing protein misfolding energetics. Science, 2003. 299(5607): p. 713–6. - PubMed
-
- Maurer MS, et al., Tafamidis Treatment for Patients with Transthyretin Amyloid Cardiomyopathy. N Engl J Med, 2018. 379: p. 1007–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

