Maintenance tobramycin primarily affects untargeted bacteria in the CF sputum microbiome
- PMID: 32631930
- PMCID: PMC7875198
- DOI: 10.1136/thoraxjnl-2019-214187
Maintenance tobramycin primarily affects untargeted bacteria in the CF sputum microbiome
Erratum in
-
Correction: Maintenance tobramycin primarily affects untargeted bacteria in the CF sputum microbiome.Thorax. 2022 Mar;77(3):e1. doi: 10.1136/thoraxjnl-2019-214187corr1. Thorax. 2022. PMID: 35177541 No abstract available.
Abstract
Rationale: The most common antibiotic used to treat people with cystic fibrosis (PWCF) is inhaled tobramycin, administered as maintenance therapy for chronic Pseudomonas aeruginosa lung infections. While the effects of inhaled tobramycin on P. aeruginosa abundance and lung function diminish with continued therapy, this maintenance treatment is known to improve long-term outcomes, underscoring how little is known about why antibiotics work in CF infections, what their effects are on complex CF sputum microbiomes and how to improve these treatments.
Objectives: To rigorously define the effect of maintenance tobramycin on CF sputum microbiome characteristics.
Methods and measurements: We collected sputum from 30 PWCF at standardised times before, during and after a single month-long course of maintenance inhaled tobramycin. We used traditional culture, quantitative PCR and metagenomic sequencing to define the dynamic effects of this treatment on sputum microbiomes, including abundance changes in both clinically targeted and untargeted bacteria, as well as functional gene categories.
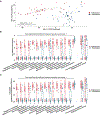
Main results: CF sputum microbiota changed most markedly by 1 week of antibiotic therapy and plateaued thereafter, and this shift was largely driven by changes in non-dominant taxa. The genetically conferred functional capacities (ie, metagenomes) of subjects' sputum communities changed little with antibiotic perturbation, despite taxonomic shifts, suggesting functional redundancy within the CF sputum microbiome.
Conclusions: Maintenance treatment with inhaled tobramycin, an antibiotic with demonstrated long-term mortality benefit, primarily impacted clinically untargeted bacteria in CF sputum, highlighting the importance of monitoring the non-canonical effects of antibiotics and other treatments to accurately define and improve their clinical impact.
Keywords: bacterial infection; cystic fibrosis; respiratory infection.
© Author(s) (or their employer(s)) 2020. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




Comment in
-
Airway microbiome studies challenge simplistic models of inhaled tobramycin benefit.Thorax. 2020 Dec;75(12):1031-1032. doi: 10.1136/thoraxjnl-2020-216005. Epub 2020 Nov 2. Thorax. 2020. PMID: 33139450 No abstract available.
References
-
- Konstan MW, Morgan WJ, Butler SM, et al. Risk factors for rate of decline in forced expiratory volume in one second in children and adolescents with cystic fibrosis. J Pediatr 2007;151:134–9. - PubMed
-
- Sanders DB, Bittner RCL, Rosenfeld M, et al. Pulmonary exacerbations are associated with subsequent FEV1 decline in both adults and children with cystic fibrosis. Pediatr Pulmonol 2011;46:393–400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
